
Suid alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
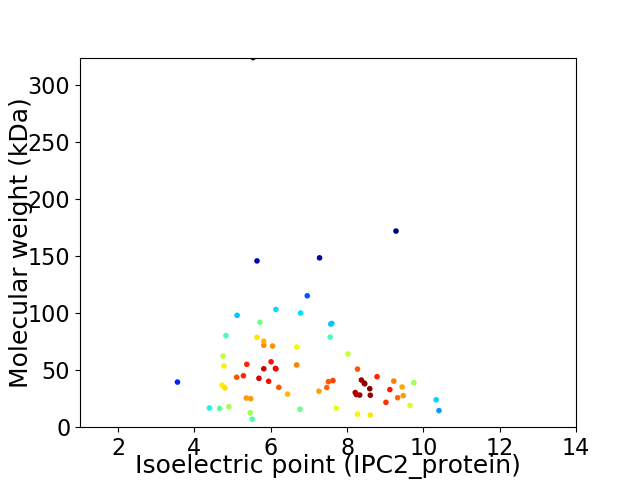
Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5PP75|Q5PP75_9ALPH Immediate-early protein IE180 OS=Suid alphaherpesvirus 1 OX=10345 GN=IE180(TRS) PE=3 SV=1
MM1 pKa = 7.59DD2 pKa = 5.22RR3 pKa = 11.84VWADD7 pKa = 2.66WYY9 pKa = 10.38EE10 pKa = 4.25PVPSPPFSPVDD21 pKa = 3.55PPGPRR26 pKa = 11.84PTTPVPGSSPPSPASTPTPPKK47 pKa = 8.29RR48 pKa = 11.84GRR50 pKa = 11.84YY51 pKa = 7.89VVEE54 pKa = 4.11HH55 pKa = 7.06PEE57 pKa = 4.1YY58 pKa = 10.82GPPPDD63 pKa = 3.94PEE65 pKa = 4.09EE66 pKa = 4.2VRR68 pKa = 11.84VHH70 pKa = 6.28GARR73 pKa = 11.84GPGAFCAAPWRR84 pKa = 11.84PDD86 pKa = 3.04TRR88 pKa = 11.84RR89 pKa = 11.84LGADD93 pKa = 3.12VNRR96 pKa = 11.84LFRR99 pKa = 11.84GIAVSAADD107 pKa = 3.48VTGDD111 pKa = 3.15TRR113 pKa = 11.84ALRR116 pKa = 11.84RR117 pKa = 11.84ALFDD121 pKa = 4.26FYY123 pKa = 11.86AMGYY127 pKa = 6.58TRR129 pKa = 11.84QRR131 pKa = 11.84PSAPCWQALLQLSPEE146 pKa = 3.76QSAPLRR152 pKa = 11.84SALRR156 pKa = 11.84EE157 pKa = 3.93LNEE160 pKa = 3.95RR161 pKa = 11.84DD162 pKa = 3.66VYY164 pKa = 10.87DD165 pKa = 4.16PRR167 pKa = 11.84VLSPPVIEE175 pKa = 4.94GPLFGEE181 pKa = 4.35EE182 pKa = 4.28CDD184 pKa = 4.4VDD186 pKa = 4.21EE187 pKa = 6.2DD188 pKa = 5.04DD189 pKa = 5.38AGSDD193 pKa = 3.3TTVASEE199 pKa = 3.76FSFRR203 pKa = 11.84GSVCEE208 pKa = 5.61DD209 pKa = 3.48DD210 pKa = 6.26GEE212 pKa = 5.66DD213 pKa = 3.76EE214 pKa = 5.57DD215 pKa = 4.98EE216 pKa = 4.74EE217 pKa = 4.59EE218 pKa = 4.97DD219 pKa = 5.04GEE221 pKa = 4.86EE222 pKa = 3.99EE223 pKa = 5.22DD224 pKa = 3.89EE225 pKa = 4.63DD226 pKa = 4.38EE227 pKa = 4.51EE228 pKa = 5.48GEE230 pKa = 4.16EE231 pKa = 4.26EE232 pKa = 3.96EE233 pKa = 5.72DD234 pKa = 3.77EE235 pKa = 4.33EE236 pKa = 4.92EE237 pKa = 4.78EE238 pKa = 4.62EE239 pKa = 4.39GDD241 pKa = 3.53EE242 pKa = 5.3DD243 pKa = 5.58GEE245 pKa = 4.35TDD247 pKa = 3.84VYY249 pKa = 11.45EE250 pKa = 4.72EE251 pKa = 4.71DD252 pKa = 5.59DD253 pKa = 3.79EE254 pKa = 7.03AEE256 pKa = 4.27DD257 pKa = 5.23EE258 pKa = 4.18EE259 pKa = 5.7DD260 pKa = 5.22EE261 pKa = 4.88EE262 pKa = 6.68DD263 pKa = 5.17GDD265 pKa = 4.55DD266 pKa = 4.41FDD268 pKa = 4.74GASVGDD274 pKa = 4.22DD275 pKa = 3.59DD276 pKa = 5.72VFEE279 pKa = 4.61PPEE282 pKa = 4.86DD283 pKa = 4.11GSDD286 pKa = 3.99GEE288 pKa = 5.04GSGSDD293 pKa = 3.8DD294 pKa = 4.01GGDD297 pKa = 4.28GEE299 pKa = 6.4DD300 pKa = 3.72EE301 pKa = 5.91DD302 pKa = 4.83EE303 pKa = 6.67DD304 pKa = 4.12EE305 pKa = 6.69DD306 pKa = 4.69EE307 pKa = 6.73DD308 pKa = 4.48EE309 pKa = 6.34DD310 pKa = 4.7EE311 pKa = 6.26DD312 pKa = 5.53DD313 pKa = 6.14GEE315 pKa = 4.74DD316 pKa = 3.7EE317 pKa = 4.2EE318 pKa = 6.18DD319 pKa = 4.28EE320 pKa = 4.28EE321 pKa = 6.54GEE323 pKa = 4.64DD324 pKa = 3.99GGEE327 pKa = 4.13DD328 pKa = 3.96GEE330 pKa = 5.6DD331 pKa = 3.73GEE333 pKa = 4.74EE334 pKa = 5.4DD335 pKa = 3.54EE336 pKa = 6.37DD337 pKa = 4.29EE338 pKa = 5.0DD339 pKa = 5.64GEE341 pKa = 4.51GEE343 pKa = 4.44EE344 pKa = 4.69GGKK347 pKa = 10.07DD348 pKa = 2.88AARR351 pKa = 11.84RR352 pKa = 11.84GTRR355 pKa = 11.84APTRR359 pKa = 11.84PAAAPP364 pKa = 3.5
MM1 pKa = 7.59DD2 pKa = 5.22RR3 pKa = 11.84VWADD7 pKa = 2.66WYY9 pKa = 10.38EE10 pKa = 4.25PVPSPPFSPVDD21 pKa = 3.55PPGPRR26 pKa = 11.84PTTPVPGSSPPSPASTPTPPKK47 pKa = 8.29RR48 pKa = 11.84GRR50 pKa = 11.84YY51 pKa = 7.89VVEE54 pKa = 4.11HH55 pKa = 7.06PEE57 pKa = 4.1YY58 pKa = 10.82GPPPDD63 pKa = 3.94PEE65 pKa = 4.09EE66 pKa = 4.2VRR68 pKa = 11.84VHH70 pKa = 6.28GARR73 pKa = 11.84GPGAFCAAPWRR84 pKa = 11.84PDD86 pKa = 3.04TRR88 pKa = 11.84RR89 pKa = 11.84LGADD93 pKa = 3.12VNRR96 pKa = 11.84LFRR99 pKa = 11.84GIAVSAADD107 pKa = 3.48VTGDD111 pKa = 3.15TRR113 pKa = 11.84ALRR116 pKa = 11.84RR117 pKa = 11.84ALFDD121 pKa = 4.26FYY123 pKa = 11.86AMGYY127 pKa = 6.58TRR129 pKa = 11.84QRR131 pKa = 11.84PSAPCWQALLQLSPEE146 pKa = 3.76QSAPLRR152 pKa = 11.84SALRR156 pKa = 11.84EE157 pKa = 3.93LNEE160 pKa = 3.95RR161 pKa = 11.84DD162 pKa = 3.66VYY164 pKa = 10.87DD165 pKa = 4.16PRR167 pKa = 11.84VLSPPVIEE175 pKa = 4.94GPLFGEE181 pKa = 4.35EE182 pKa = 4.28CDD184 pKa = 4.4VDD186 pKa = 4.21EE187 pKa = 6.2DD188 pKa = 5.04DD189 pKa = 5.38AGSDD193 pKa = 3.3TTVASEE199 pKa = 3.76FSFRR203 pKa = 11.84GSVCEE208 pKa = 5.61DD209 pKa = 3.48DD210 pKa = 6.26GEE212 pKa = 5.66DD213 pKa = 3.76EE214 pKa = 5.57DD215 pKa = 4.98EE216 pKa = 4.74EE217 pKa = 4.59EE218 pKa = 4.97DD219 pKa = 5.04GEE221 pKa = 4.86EE222 pKa = 3.99EE223 pKa = 5.22DD224 pKa = 3.89EE225 pKa = 4.63DD226 pKa = 4.38EE227 pKa = 4.51EE228 pKa = 5.48GEE230 pKa = 4.16EE231 pKa = 4.26EE232 pKa = 3.96EE233 pKa = 5.72DD234 pKa = 3.77EE235 pKa = 4.33EE236 pKa = 4.92EE237 pKa = 4.78EE238 pKa = 4.62EE239 pKa = 4.39GDD241 pKa = 3.53EE242 pKa = 5.3DD243 pKa = 5.58GEE245 pKa = 4.35TDD247 pKa = 3.84VYY249 pKa = 11.45EE250 pKa = 4.72EE251 pKa = 4.71DD252 pKa = 5.59DD253 pKa = 3.79EE254 pKa = 7.03AEE256 pKa = 4.27DD257 pKa = 5.23EE258 pKa = 4.18EE259 pKa = 5.7DD260 pKa = 5.22EE261 pKa = 4.88EE262 pKa = 6.68DD263 pKa = 5.17GDD265 pKa = 4.55DD266 pKa = 4.41FDD268 pKa = 4.74GASVGDD274 pKa = 4.22DD275 pKa = 3.59DD276 pKa = 5.72VFEE279 pKa = 4.61PPEE282 pKa = 4.86DD283 pKa = 4.11GSDD286 pKa = 3.99GEE288 pKa = 5.04GSGSDD293 pKa = 3.8DD294 pKa = 4.01GGDD297 pKa = 4.28GEE299 pKa = 6.4DD300 pKa = 3.72EE301 pKa = 5.91DD302 pKa = 4.83EE303 pKa = 6.67DD304 pKa = 4.12EE305 pKa = 6.69DD306 pKa = 4.69EE307 pKa = 6.73DD308 pKa = 4.48EE309 pKa = 6.34DD310 pKa = 4.7EE311 pKa = 6.26DD312 pKa = 5.53DD313 pKa = 6.14GEE315 pKa = 4.74DD316 pKa = 3.7EE317 pKa = 4.2EE318 pKa = 6.18DD319 pKa = 4.28EE320 pKa = 4.28EE321 pKa = 6.54GEE323 pKa = 4.64DD324 pKa = 3.99GGEE327 pKa = 4.13DD328 pKa = 3.96GEE330 pKa = 5.6DD331 pKa = 3.73GEE333 pKa = 4.74EE334 pKa = 5.4DD335 pKa = 3.54EE336 pKa = 6.37DD337 pKa = 4.29EE338 pKa = 5.0DD339 pKa = 5.64GEE341 pKa = 4.51GEE343 pKa = 4.44EE344 pKa = 4.69GGKK347 pKa = 10.07DD348 pKa = 2.88AARR351 pKa = 11.84RR352 pKa = 11.84GTRR355 pKa = 11.84APTRR359 pKa = 11.84PAAAPP364 pKa = 3.5
Molecular weight: 39.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G3G8W3|G3G8W3_9ALPH DNA primase OS=Suid alphaherpesvirus 1 OX=10345 GN=UL52 PE=3 SV=1
MM1 pKa = 7.81EE2 pKa = 6.23DD3 pKa = 3.58SGNSSGSEE11 pKa = 3.65ASRR14 pKa = 11.84SGSEE18 pKa = 3.41EE19 pKa = 3.47RR20 pKa = 11.84RR21 pKa = 11.84PVRR24 pKa = 11.84EE25 pKa = 3.98RR26 pKa = 11.84LGSRR30 pKa = 11.84PPEE33 pKa = 4.01RR34 pKa = 11.84RR35 pKa = 11.84PVRR38 pKa = 11.84ARR40 pKa = 11.84LGAIRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGRR51 pKa = 11.84GGRR54 pKa = 11.84AARR57 pKa = 11.84QALRR61 pKa = 11.84QRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84QQQQQQRR72 pKa = 11.84QQQHH76 pKa = 3.04QRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84QEE82 pKa = 3.26ADD84 pKa = 3.57RR85 pKa = 11.84PDD87 pKa = 4.37GGPDD91 pKa = 3.32APPDD95 pKa = 3.74RR96 pKa = 11.84LSEE99 pKa = 4.18SARR102 pKa = 11.84AAVSATHH109 pKa = 6.17ARR111 pKa = 11.84VGATRR116 pKa = 11.84VNEE119 pKa = 4.05LFASARR125 pKa = 11.84HH126 pKa = 5.78DD127 pKa = 3.5LSRR130 pKa = 11.84PVFNDD135 pKa = 3.39GFRR138 pKa = 11.84AAGSSPWAAVLEE150 pKa = 4.87FGAEE154 pKa = 3.86QFTPDD159 pKa = 3.16GRR161 pKa = 11.84RR162 pKa = 11.84VTWEE166 pKa = 3.44TLMFHH171 pKa = 6.9GADD174 pKa = 3.25LHH176 pKa = 6.84RR177 pKa = 11.84LFEE180 pKa = 4.36VRR182 pKa = 11.84PHH184 pKa = 5.21ATEE187 pKa = 3.54AARR190 pKa = 11.84VLRR193 pKa = 11.84EE194 pKa = 3.8MVLLNEE200 pKa = 4.7GLTEE204 pKa = 4.17SLASADD210 pKa = 4.06EE211 pKa = 4.03TLTWVKK217 pKa = 10.87LILTKK222 pKa = 10.87GLTLRR227 pKa = 11.84TLDD230 pKa = 4.98PIVATAGAVLQNLRR244 pKa = 11.84LKK246 pKa = 10.67LGPFLRR252 pKa = 11.84CYY254 pKa = 10.85LRR256 pKa = 11.84DD257 pKa = 3.65TPVDD261 pKa = 3.25EE262 pKa = 4.21LVRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84LRR270 pKa = 11.84DD271 pKa = 3.01VRR273 pKa = 11.84CIVTYY278 pKa = 8.89TLVMLARR285 pKa = 11.84IARR288 pKa = 11.84VVEE291 pKa = 4.35RR292 pKa = 11.84GSSCVLPEE300 pKa = 4.69DD301 pKa = 5.18LGDD304 pKa = 3.79SPVPLEE310 pKa = 4.14EE311 pKa = 4.26YY312 pKa = 11.14VPGACLGGIMDD323 pKa = 5.53ALDD326 pKa = 3.55SHH328 pKa = 6.5KK329 pKa = 9.4TGCDD333 pKa = 3.09APTCRR338 pKa = 11.84LTCSYY343 pKa = 10.28TLVPVYY349 pKa = 9.35MHH351 pKa = 6.9GKK353 pKa = 9.54YY354 pKa = 10.31FYY356 pKa = 10.45CNHH359 pKa = 6.76LFF361 pKa = 3.62
MM1 pKa = 7.81EE2 pKa = 6.23DD3 pKa = 3.58SGNSSGSEE11 pKa = 3.65ASRR14 pKa = 11.84SGSEE18 pKa = 3.41EE19 pKa = 3.47RR20 pKa = 11.84RR21 pKa = 11.84PVRR24 pKa = 11.84EE25 pKa = 3.98RR26 pKa = 11.84LGSRR30 pKa = 11.84PPEE33 pKa = 4.01RR34 pKa = 11.84RR35 pKa = 11.84PVRR38 pKa = 11.84ARR40 pKa = 11.84LGAIRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGRR51 pKa = 11.84GGRR54 pKa = 11.84AARR57 pKa = 11.84QALRR61 pKa = 11.84QRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84QQQQQQRR72 pKa = 11.84QQQHH76 pKa = 3.04QRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84QEE82 pKa = 3.26ADD84 pKa = 3.57RR85 pKa = 11.84PDD87 pKa = 4.37GGPDD91 pKa = 3.32APPDD95 pKa = 3.74RR96 pKa = 11.84LSEE99 pKa = 4.18SARR102 pKa = 11.84AAVSATHH109 pKa = 6.17ARR111 pKa = 11.84VGATRR116 pKa = 11.84VNEE119 pKa = 4.05LFASARR125 pKa = 11.84HH126 pKa = 5.78DD127 pKa = 3.5LSRR130 pKa = 11.84PVFNDD135 pKa = 3.39GFRR138 pKa = 11.84AAGSSPWAAVLEE150 pKa = 4.87FGAEE154 pKa = 3.86QFTPDD159 pKa = 3.16GRR161 pKa = 11.84RR162 pKa = 11.84VTWEE166 pKa = 3.44TLMFHH171 pKa = 6.9GADD174 pKa = 3.25LHH176 pKa = 6.84RR177 pKa = 11.84LFEE180 pKa = 4.36VRR182 pKa = 11.84PHH184 pKa = 5.21ATEE187 pKa = 3.54AARR190 pKa = 11.84VLRR193 pKa = 11.84EE194 pKa = 3.8MVLLNEE200 pKa = 4.7GLTEE204 pKa = 4.17SLASADD210 pKa = 4.06EE211 pKa = 4.03TLTWVKK217 pKa = 10.87LILTKK222 pKa = 10.87GLTLRR227 pKa = 11.84TLDD230 pKa = 4.98PIVATAGAVLQNLRR244 pKa = 11.84LKK246 pKa = 10.67LGPFLRR252 pKa = 11.84CYY254 pKa = 10.85LRR256 pKa = 11.84DD257 pKa = 3.65TPVDD261 pKa = 3.25EE262 pKa = 4.21LVRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84LRR270 pKa = 11.84DD271 pKa = 3.01VRR273 pKa = 11.84CIVTYY278 pKa = 8.89TLVMLARR285 pKa = 11.84IARR288 pKa = 11.84VVEE291 pKa = 4.35RR292 pKa = 11.84GSSCVLPEE300 pKa = 4.69DD301 pKa = 5.18LGDD304 pKa = 3.79SPVPLEE310 pKa = 4.14EE311 pKa = 4.26YY312 pKa = 11.14VPGACLGGIMDD323 pKa = 5.53ALDD326 pKa = 3.55SHH328 pKa = 6.5KK329 pKa = 9.4TGCDD333 pKa = 3.09APTCRR338 pKa = 11.84LTCSYY343 pKa = 10.28TLVPVYY349 pKa = 9.35MHH351 pKa = 6.9GKK353 pKa = 9.54YY354 pKa = 10.31FYY356 pKa = 10.45CNHH359 pKa = 6.76LFF361 pKa = 3.62
Molecular weight: 40.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
35751 |
63 |
3084 |
503.5 |
53.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.614 ± 0.533 | 1.589 ± 0.121 |
5.645 ± 0.203 | 5.77 ± 0.232 |
3.217 ± 0.202 | 8.492 ± 0.772 |
2.447 ± 0.155 | 1.885 ± 0.144 |
1.236 ± 0.097 | 9.818 ± 0.289 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.592 ± 0.126 | 1.698 ± 0.152 |
8.341 ± 0.546 | 2.45 ± 0.143 |
9.656 ± 0.221 | 5.046 ± 0.294 |
4.696 ± 0.211 | 7.544 ± 0.256 |
0.962 ± 0.066 | 2.302 ± 0.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
