
Hubei odonate virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
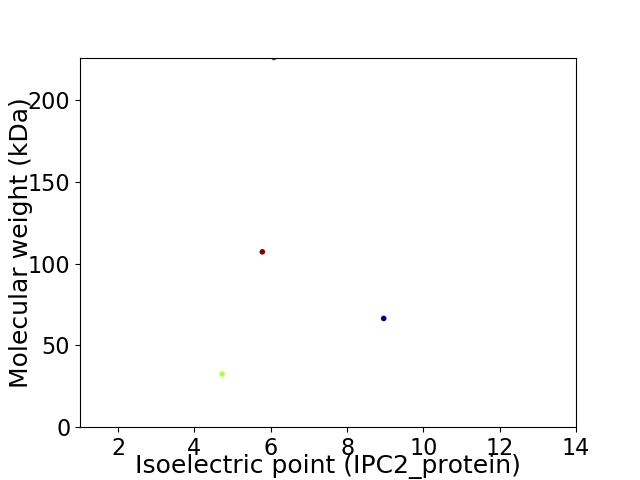
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
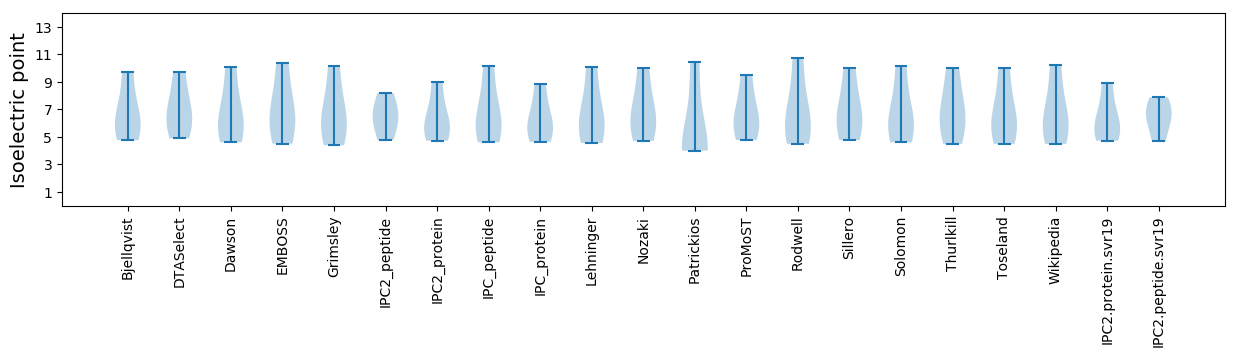
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KKX6|A0A1L3KKX6_9VIRU Uncharacterized protein OS=Hubei odonate virus 6 OX=1923001 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 5.56LLAEE6 pKa = 4.01VWEE9 pKa = 4.54RR10 pKa = 11.84DD11 pKa = 3.37TTLVDD16 pKa = 3.47VLDD19 pKa = 4.61DD20 pKa = 3.96SGIGEE25 pKa = 4.19IPSIQSKK32 pKa = 10.4INAIQEE38 pKa = 4.14EE39 pKa = 4.39QSQQKK44 pKa = 10.23DD45 pKa = 3.34QLDD48 pKa = 4.28SLGHH52 pKa = 5.44TVDD55 pKa = 4.54NNQKK59 pKa = 10.21SNEE62 pKa = 3.71ARR64 pKa = 11.84ILDD67 pKa = 4.64LYY69 pKa = 10.93TNVNSNTRR77 pKa = 11.84DD78 pKa = 3.54LQRR81 pKa = 11.84LEE83 pKa = 4.67QEE85 pKa = 4.57VKK87 pKa = 10.83DD88 pKa = 3.79LTLSIGKK95 pKa = 8.54IQLQMHH101 pKa = 5.93EE102 pKa = 5.02LEE104 pKa = 5.07LKK106 pKa = 10.58VSSNSQIIDD115 pKa = 3.4DD116 pKa = 3.9MTYY119 pKa = 10.95DD120 pKa = 3.78LEE122 pKa = 4.54VVKK125 pKa = 11.16GNVLTLAQDD134 pKa = 3.52MNKK137 pKa = 10.28LNNEE141 pKa = 4.12LLLVKK146 pKa = 10.24PDD148 pKa = 3.39VSEE151 pKa = 4.95LKK153 pKa = 9.91SQNVFTYY160 pKa = 9.78PFSVYY165 pKa = 11.19ANIFLQDD172 pKa = 2.82ICAVYY177 pKa = 10.08EE178 pKa = 4.28GSKK181 pKa = 8.68VTIVDD186 pKa = 3.07IGGSSIEE193 pKa = 4.18YY194 pKa = 7.66QTTTIGSGMGRR205 pKa = 11.84KK206 pKa = 9.12VSSPVKK212 pKa = 8.84YY213 pKa = 9.34VKK215 pKa = 10.19RR216 pKa = 11.84YY217 pKa = 8.27EE218 pKa = 4.09LVPSVVFNKK227 pKa = 10.02KK228 pKa = 8.15VYY230 pKa = 10.28QYY232 pKa = 11.11ISFVGINRR240 pKa = 11.84TTSQPAFSDD249 pKa = 3.83DD250 pKa = 4.88PIVTWGYY257 pKa = 9.37QVIKK261 pKa = 10.63LSNNVNLDD269 pKa = 3.01AMSIMLIKK277 pKa = 9.98EE278 pKa = 3.8AYY280 pKa = 9.87VPVNFSKK287 pKa = 10.69IKK289 pKa = 10.33
MM1 pKa = 7.99DD2 pKa = 5.56LLAEE6 pKa = 4.01VWEE9 pKa = 4.54RR10 pKa = 11.84DD11 pKa = 3.37TTLVDD16 pKa = 3.47VLDD19 pKa = 4.61DD20 pKa = 3.96SGIGEE25 pKa = 4.19IPSIQSKK32 pKa = 10.4INAIQEE38 pKa = 4.14EE39 pKa = 4.39QSQQKK44 pKa = 10.23DD45 pKa = 3.34QLDD48 pKa = 4.28SLGHH52 pKa = 5.44TVDD55 pKa = 4.54NNQKK59 pKa = 10.21SNEE62 pKa = 3.71ARR64 pKa = 11.84ILDD67 pKa = 4.64LYY69 pKa = 10.93TNVNSNTRR77 pKa = 11.84DD78 pKa = 3.54LQRR81 pKa = 11.84LEE83 pKa = 4.67QEE85 pKa = 4.57VKK87 pKa = 10.83DD88 pKa = 3.79LTLSIGKK95 pKa = 8.54IQLQMHH101 pKa = 5.93EE102 pKa = 5.02LEE104 pKa = 5.07LKK106 pKa = 10.58VSSNSQIIDD115 pKa = 3.4DD116 pKa = 3.9MTYY119 pKa = 10.95DD120 pKa = 3.78LEE122 pKa = 4.54VVKK125 pKa = 11.16GNVLTLAQDD134 pKa = 3.52MNKK137 pKa = 10.28LNNEE141 pKa = 4.12LLLVKK146 pKa = 10.24PDD148 pKa = 3.39VSEE151 pKa = 4.95LKK153 pKa = 9.91SQNVFTYY160 pKa = 9.78PFSVYY165 pKa = 11.19ANIFLQDD172 pKa = 2.82ICAVYY177 pKa = 10.08EE178 pKa = 4.28GSKK181 pKa = 8.68VTIVDD186 pKa = 3.07IGGSSIEE193 pKa = 4.18YY194 pKa = 7.66QTTTIGSGMGRR205 pKa = 11.84KK206 pKa = 9.12VSSPVKK212 pKa = 8.84YY213 pKa = 9.34VKK215 pKa = 10.19RR216 pKa = 11.84YY217 pKa = 8.27EE218 pKa = 4.09LVPSVVFNKK227 pKa = 10.02KK228 pKa = 8.15VYY230 pKa = 10.28QYY232 pKa = 11.11ISFVGINRR240 pKa = 11.84TTSQPAFSDD249 pKa = 3.83DD250 pKa = 4.88PIVTWGYY257 pKa = 9.37QVIKK261 pKa = 10.63LSNNVNLDD269 pKa = 3.01AMSIMLIKK277 pKa = 9.98EE278 pKa = 3.8AYY280 pKa = 9.87VPVNFSKK287 pKa = 10.69IKK289 pKa = 10.33
Molecular weight: 32.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKV8|A0A1L3KKV8_9VIRU Uncharacterized protein OS=Hubei odonate virus 6 OX=1923001 PE=4 SV=1
MM1 pKa = 6.98TTRR4 pKa = 11.84KK5 pKa = 9.9CDD7 pKa = 4.4DD8 pKa = 3.51ILRR11 pKa = 11.84KK12 pKa = 9.37EE13 pKa = 4.27PNYY16 pKa = 10.58SRR18 pKa = 11.84QNTTLANDD26 pKa = 3.26INIDD30 pKa = 3.46LKK32 pKa = 10.25RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.84RR36 pKa = 11.84ISPYY40 pKa = 10.67LLDD43 pKa = 3.92MLNEE47 pKa = 4.2PVISDD52 pKa = 4.31KK53 pKa = 10.72PVKK56 pKa = 8.91KK57 pKa = 10.53CKK59 pKa = 7.95VARR62 pKa = 11.84MTGHH66 pKa = 5.39TTRR69 pKa = 11.84KK70 pKa = 9.54YY71 pKa = 8.61STKK74 pKa = 10.13TRR76 pKa = 11.84DD77 pKa = 3.64LNSKK81 pKa = 6.71SHH83 pKa = 6.55KK84 pKa = 10.4VMTVTRR90 pKa = 11.84PEE92 pKa = 3.81NRR94 pKa = 11.84QVTVIQKK101 pKa = 9.69VEE103 pKa = 4.02EE104 pKa = 4.17VQPKK108 pKa = 8.15LTCKK112 pKa = 9.07QRR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.07MRR118 pKa = 11.84SSLRR122 pKa = 11.84ICLYY126 pKa = 10.92NKK128 pKa = 9.8ANKK131 pKa = 10.16GKK133 pKa = 8.0TLLEE137 pKa = 4.04NKK139 pKa = 8.63TWVEE143 pKa = 4.26KK144 pKa = 10.57IAISDD149 pKa = 3.49RR150 pKa = 11.84DD151 pKa = 3.74QVPEE155 pKa = 5.48AIQQCYY161 pKa = 9.5IPPPRR166 pKa = 11.84KK167 pKa = 9.17QSYY170 pKa = 8.65EE171 pKa = 4.06RR172 pKa = 11.84IVPGRR177 pKa = 11.84VNLNTIDD184 pKa = 4.66YY185 pKa = 7.76EE186 pKa = 4.65VHH188 pKa = 7.32DD189 pKa = 4.43NWTKK193 pKa = 11.06LSDD196 pKa = 3.36QDD198 pKa = 3.97VGVKK202 pKa = 9.76FSKK205 pKa = 10.66RR206 pKa = 11.84MTHH209 pKa = 6.61NSDD212 pKa = 3.14EE213 pKa = 4.31NALSKK218 pKa = 10.92LFAEE222 pKa = 4.32IRR224 pKa = 11.84KK225 pKa = 8.78EE226 pKa = 3.91FSEE229 pKa = 4.53TEE231 pKa = 3.94LEE233 pKa = 4.19RR234 pKa = 11.84KK235 pKa = 9.54VRR237 pKa = 11.84EE238 pKa = 3.81QVKK241 pKa = 10.6DD242 pKa = 3.48VTQTLLSTQNATVDD256 pKa = 3.59RR257 pKa = 11.84LIKK260 pKa = 10.26EE261 pKa = 4.2VVALRR266 pKa = 11.84EE267 pKa = 3.92EE268 pKa = 4.42LNRR271 pKa = 11.84ANAEE275 pKa = 4.11VASLKK280 pKa = 10.53RR281 pKa = 11.84KK282 pKa = 10.28LEE284 pKa = 4.14VIPEE288 pKa = 4.02VEE290 pKa = 4.33QQCSVVVPSTTIPTRR305 pKa = 11.84LLPLKK310 pKa = 10.19KK311 pKa = 10.25RR312 pKa = 11.84IIAEE316 pKa = 3.85VSKK319 pKa = 11.28ADD321 pKa = 3.5TTIVSEE327 pKa = 4.25PEE329 pKa = 3.79KK330 pKa = 10.76VNPVIKK336 pKa = 10.43KK337 pKa = 6.63VTPKK341 pKa = 10.75VIVSPKK347 pKa = 9.63IDD349 pKa = 3.09VKK351 pKa = 11.61ANVQIDD357 pKa = 3.86DD358 pKa = 4.62EE359 pKa = 4.81KK360 pKa = 11.24PSTSKK365 pKa = 10.51QALIKK370 pKa = 9.76TNSEE374 pKa = 4.09VASEE378 pKa = 3.94IAKK381 pKa = 8.76VQKK384 pKa = 10.14PSEE387 pKa = 3.96IVIKK391 pKa = 10.57QKK393 pKa = 11.12SDD395 pKa = 3.16LTFEE399 pKa = 4.3SRR401 pKa = 11.84VIPKK405 pKa = 10.02IKK407 pKa = 9.2EE408 pKa = 4.15TYY410 pKa = 8.01TDD412 pKa = 3.65RR413 pKa = 11.84DD414 pKa = 2.76IDD416 pKa = 3.41MWIDD420 pKa = 3.62FNLKK424 pKa = 10.42NEE426 pKa = 4.18DD427 pKa = 3.44VRR429 pKa = 11.84KK430 pKa = 10.35KK431 pKa = 10.66EE432 pKa = 4.01ILDD435 pKa = 3.77SLIEE439 pKa = 4.48DD440 pKa = 4.15DD441 pKa = 5.8KK442 pKa = 12.0SFASLFDD449 pKa = 3.4KK450 pKa = 10.73HH451 pKa = 6.86ASPTQRR457 pKa = 11.84LNFAKK462 pKa = 10.3LGCEE466 pKa = 3.95SSRR469 pKa = 11.84ARR471 pKa = 11.84INAQDD476 pKa = 3.38INRR479 pKa = 11.84FTWTKK484 pKa = 9.92ICAKK488 pKa = 9.84HH489 pKa = 5.45KK490 pKa = 9.4TDD492 pKa = 3.61AEE494 pKa = 4.2RR495 pKa = 11.84KK496 pKa = 8.96VEE498 pKa = 3.96CDD500 pKa = 3.48KK501 pKa = 11.14IRR503 pKa = 11.84ATLFRR508 pKa = 11.84TLFFRR513 pKa = 11.84KK514 pKa = 8.88QAEE517 pKa = 4.35FKK519 pKa = 10.54KK520 pKa = 10.95LNLGVVNPCLIGVNRR535 pKa = 11.84WITKK539 pKa = 9.84LLSLKK544 pKa = 9.16TAEE547 pKa = 3.57EE548 pKa = 4.17RR549 pKa = 11.84IYY551 pKa = 10.98FRR553 pKa = 11.84TSWNNKK559 pKa = 7.29VDD561 pKa = 4.47AITIRR566 pKa = 11.84LQNLAKK572 pKa = 10.24QKK574 pKa = 10.7CC575 pKa = 3.45
MM1 pKa = 6.98TTRR4 pKa = 11.84KK5 pKa = 9.9CDD7 pKa = 4.4DD8 pKa = 3.51ILRR11 pKa = 11.84KK12 pKa = 9.37EE13 pKa = 4.27PNYY16 pKa = 10.58SRR18 pKa = 11.84QNTTLANDD26 pKa = 3.26INIDD30 pKa = 3.46LKK32 pKa = 10.25RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.84RR36 pKa = 11.84ISPYY40 pKa = 10.67LLDD43 pKa = 3.92MLNEE47 pKa = 4.2PVISDD52 pKa = 4.31KK53 pKa = 10.72PVKK56 pKa = 8.91KK57 pKa = 10.53CKK59 pKa = 7.95VARR62 pKa = 11.84MTGHH66 pKa = 5.39TTRR69 pKa = 11.84KK70 pKa = 9.54YY71 pKa = 8.61STKK74 pKa = 10.13TRR76 pKa = 11.84DD77 pKa = 3.64LNSKK81 pKa = 6.71SHH83 pKa = 6.55KK84 pKa = 10.4VMTVTRR90 pKa = 11.84PEE92 pKa = 3.81NRR94 pKa = 11.84QVTVIQKK101 pKa = 9.69VEE103 pKa = 4.02EE104 pKa = 4.17VQPKK108 pKa = 8.15LTCKK112 pKa = 9.07QRR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.07MRR118 pKa = 11.84SSLRR122 pKa = 11.84ICLYY126 pKa = 10.92NKK128 pKa = 9.8ANKK131 pKa = 10.16GKK133 pKa = 8.0TLLEE137 pKa = 4.04NKK139 pKa = 8.63TWVEE143 pKa = 4.26KK144 pKa = 10.57IAISDD149 pKa = 3.49RR150 pKa = 11.84DD151 pKa = 3.74QVPEE155 pKa = 5.48AIQQCYY161 pKa = 9.5IPPPRR166 pKa = 11.84KK167 pKa = 9.17QSYY170 pKa = 8.65EE171 pKa = 4.06RR172 pKa = 11.84IVPGRR177 pKa = 11.84VNLNTIDD184 pKa = 4.66YY185 pKa = 7.76EE186 pKa = 4.65VHH188 pKa = 7.32DD189 pKa = 4.43NWTKK193 pKa = 11.06LSDD196 pKa = 3.36QDD198 pKa = 3.97VGVKK202 pKa = 9.76FSKK205 pKa = 10.66RR206 pKa = 11.84MTHH209 pKa = 6.61NSDD212 pKa = 3.14EE213 pKa = 4.31NALSKK218 pKa = 10.92LFAEE222 pKa = 4.32IRR224 pKa = 11.84KK225 pKa = 8.78EE226 pKa = 3.91FSEE229 pKa = 4.53TEE231 pKa = 3.94LEE233 pKa = 4.19RR234 pKa = 11.84KK235 pKa = 9.54VRR237 pKa = 11.84EE238 pKa = 3.81QVKK241 pKa = 10.6DD242 pKa = 3.48VTQTLLSTQNATVDD256 pKa = 3.59RR257 pKa = 11.84LIKK260 pKa = 10.26EE261 pKa = 4.2VVALRR266 pKa = 11.84EE267 pKa = 3.92EE268 pKa = 4.42LNRR271 pKa = 11.84ANAEE275 pKa = 4.11VASLKK280 pKa = 10.53RR281 pKa = 11.84KK282 pKa = 10.28LEE284 pKa = 4.14VIPEE288 pKa = 4.02VEE290 pKa = 4.33QQCSVVVPSTTIPTRR305 pKa = 11.84LLPLKK310 pKa = 10.19KK311 pKa = 10.25RR312 pKa = 11.84IIAEE316 pKa = 3.85VSKK319 pKa = 11.28ADD321 pKa = 3.5TTIVSEE327 pKa = 4.25PEE329 pKa = 3.79KK330 pKa = 10.76VNPVIKK336 pKa = 10.43KK337 pKa = 6.63VTPKK341 pKa = 10.75VIVSPKK347 pKa = 9.63IDD349 pKa = 3.09VKK351 pKa = 11.61ANVQIDD357 pKa = 3.86DD358 pKa = 4.62EE359 pKa = 4.81KK360 pKa = 11.24PSTSKK365 pKa = 10.51QALIKK370 pKa = 9.76TNSEE374 pKa = 4.09VASEE378 pKa = 3.94IAKK381 pKa = 8.76VQKK384 pKa = 10.14PSEE387 pKa = 3.96IVIKK391 pKa = 10.57QKK393 pKa = 11.12SDD395 pKa = 3.16LTFEE399 pKa = 4.3SRR401 pKa = 11.84VIPKK405 pKa = 10.02IKK407 pKa = 9.2EE408 pKa = 4.15TYY410 pKa = 8.01TDD412 pKa = 3.65RR413 pKa = 11.84DD414 pKa = 2.76IDD416 pKa = 3.41MWIDD420 pKa = 3.62FNLKK424 pKa = 10.42NEE426 pKa = 4.18DD427 pKa = 3.44VRR429 pKa = 11.84KK430 pKa = 10.35KK431 pKa = 10.66EE432 pKa = 4.01ILDD435 pKa = 3.77SLIEE439 pKa = 4.48DD440 pKa = 4.15DD441 pKa = 5.8KK442 pKa = 12.0SFASLFDD449 pKa = 3.4KK450 pKa = 10.73HH451 pKa = 6.86ASPTQRR457 pKa = 11.84LNFAKK462 pKa = 10.3LGCEE466 pKa = 3.95SSRR469 pKa = 11.84ARR471 pKa = 11.84INAQDD476 pKa = 3.38INRR479 pKa = 11.84FTWTKK484 pKa = 9.92ICAKK488 pKa = 9.84HH489 pKa = 5.45KK490 pKa = 9.4TDD492 pKa = 3.61AEE494 pKa = 4.2RR495 pKa = 11.84KK496 pKa = 8.96VEE498 pKa = 3.96CDD500 pKa = 3.48KK501 pKa = 11.14IRR503 pKa = 11.84ATLFRR508 pKa = 11.84TLFFRR513 pKa = 11.84KK514 pKa = 8.88QAEE517 pKa = 4.35FKK519 pKa = 10.54KK520 pKa = 10.95LNLGVVNPCLIGVNRR535 pKa = 11.84WITKK539 pKa = 9.84LLSLKK544 pKa = 9.16TAEE547 pKa = 3.57EE548 pKa = 4.17RR549 pKa = 11.84IYY551 pKa = 10.98FRR553 pKa = 11.84TSWNNKK559 pKa = 7.29VDD561 pKa = 4.47AITIRR566 pKa = 11.84LQNLAKK572 pKa = 10.24QKK574 pKa = 10.7CC575 pKa = 3.45
Molecular weight: 66.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3826 |
289 |
1991 |
956.5 |
108.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.829 ± 0.55 | 1.464 ± 0.194 |
5.567 ± 0.395 | 6.325 ± 0.307 |
3.999 ± 0.533 | 5.018 ± 0.99 |
1.777 ± 0.5 | 6.926 ± 0.438 |
7.188 ± 1.33 | 9.2 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.934 ± 0.164 | 5.227 ± 0.384 |
4.339 ± 0.926 | 4.182 ± 0.392 |
4.861 ± 0.732 | 7.371 ± 0.317 |
6.717 ± 0.656 | 7.527 ± 0.566 |
1.176 ± 0.138 | 3.372 ± 0.488 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
