
Roseobacter cerasinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseobacter
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
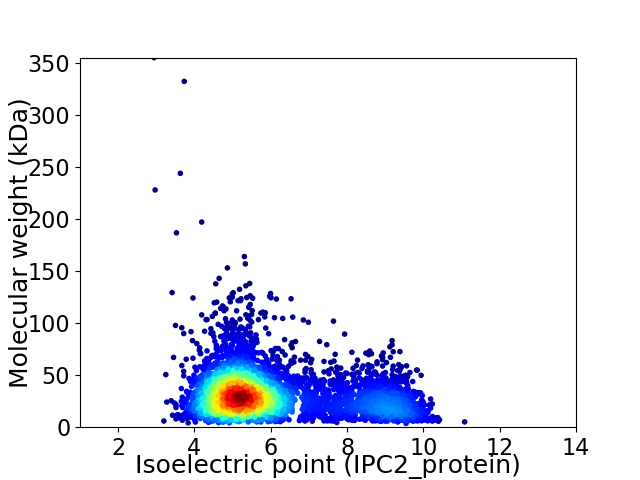
Virtual 2D-PAGE plot for 4343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A640VQK1|A0A640VQK1_9RHOB Shikimate kinase OS=Roseobacter cerasinus OX=2602289 GN=aroK_1 PE=3 SV=1
MM1 pKa = 7.48SNIDD5 pKa = 3.97PSQSSQNPEE14 pKa = 3.88TNQDD18 pKa = 3.82DD19 pKa = 4.46LSLHH23 pKa = 7.0LSTCTCAACMADD35 pKa = 6.09DD36 pKa = 4.95RR37 pKa = 11.84FQDD40 pKa = 3.58DD41 pKa = 3.98VEE43 pKa = 4.69RR44 pKa = 11.84SGNPTSGGGTAAAATGKK61 pKa = 7.26TLRR64 pKa = 11.84EE65 pKa = 3.75MADD68 pKa = 3.87FLEE71 pKa = 4.22MGYY74 pKa = 9.48WGNEE78 pKa = 3.16VGLRR82 pKa = 11.84HH83 pKa = 6.02NVGTTGLDD91 pKa = 3.41PNNGVLHH98 pKa = 5.88YY99 pKa = 9.47NVSGFGPLTYY109 pKa = 10.63GGGSDD114 pKa = 3.86ANGVSSARR122 pKa = 11.84AALIRR127 pKa = 11.84DD128 pKa = 3.45AFDD131 pKa = 2.94VYY133 pKa = 10.9EE134 pKa = 4.47AVLGIQFVEE143 pKa = 4.6TTSTNDD149 pKa = 3.14SVVDD153 pKa = 4.84FFFSDD158 pKa = 3.5NQSGAFAGSTRR169 pKa = 11.84YY170 pKa = 10.38GDD172 pKa = 3.26GTIYY176 pKa = 10.68YY177 pKa = 9.89SYY179 pKa = 11.3INIAASWSGSSSTYY193 pKa = 11.06DD194 pKa = 3.7DD195 pKa = 3.89YY196 pKa = 11.86TLQTIFHH203 pKa = 7.27EE204 pKa = 4.72IGHH207 pKa = 6.51ALGLGHH213 pKa = 6.5QGPYY217 pKa = 10.01NGSASYY223 pKa = 11.6ANDD226 pKa = 4.42AIFALDD232 pKa = 3.13SWQASMMSYY241 pKa = 10.57FSQTEE246 pKa = 3.62NSAISASYY254 pKa = 10.81EE255 pKa = 3.91FLQTPMAVDD264 pKa = 3.78WLALDD269 pKa = 4.84SIYY272 pKa = 10.8GQQGYY277 pKa = 9.76GISNAFVGDD286 pKa = 3.46TVYY289 pKa = 11.14GFNTNIAASEE299 pKa = 3.96SRR301 pKa = 11.84IWNEE305 pKa = 3.12FSAYY309 pKa = 10.06AGRR312 pKa = 11.84TASTIVDD319 pKa = 3.39GGGIDD324 pKa = 3.45TLDD327 pKa = 3.51FSGYY331 pKa = 9.78SANQKK336 pKa = 9.74IDD338 pKa = 3.67LTVQTADD345 pKa = 3.13QTSQNTSNVGGRR357 pKa = 11.84TGNLTLAIGTVIEE370 pKa = 4.1NAIGGSGNDD379 pKa = 3.24YY380 pKa = 10.94FIGNEE385 pKa = 3.63ADD387 pKa = 3.25NVFRR391 pKa = 11.84GGAGNDD397 pKa = 3.16RR398 pKa = 11.84FIGQLGDD405 pKa = 3.63DD406 pKa = 4.17TFHH409 pKa = 7.0GNAGLDD415 pKa = 3.39TVIYY419 pKa = 9.92NLVFGAYY426 pKa = 9.16SFSVLSDD433 pKa = 3.53VIQVIGEE440 pKa = 4.56GIDD443 pKa = 3.57LVYY446 pKa = 10.06DD447 pKa = 4.17TIEE450 pKa = 3.98TFQFSDD456 pKa = 2.78ITYY459 pKa = 9.64SFTDD463 pKa = 3.28IFALVGNAVPTARR476 pKa = 11.84DD477 pKa = 3.23DD478 pKa = 3.74SATVEE483 pKa = 4.12EE484 pKa = 5.26GATITLSVLDD494 pKa = 4.91NDD496 pKa = 4.55TDD498 pKa = 4.31PDD500 pKa = 4.39GDD502 pKa = 4.1PLTITEE508 pKa = 4.54VNGQAIGVNEE518 pKa = 4.25TVDD521 pKa = 3.96LASGASVTLRR531 pKa = 11.84DD532 pKa = 5.0DD533 pKa = 3.57GTLSYY538 pKa = 10.5AQNGVFDD545 pKa = 4.09TLNTGEE551 pKa = 4.34SGGDD555 pKa = 3.21SFEE558 pKa = 4.41YY559 pKa = 10.53AVSDD563 pKa = 3.99GKK565 pKa = 11.26GGAATATVVMTVEE578 pKa = 4.45GAGDD582 pKa = 4.21DD583 pKa = 4.11SPSAAIGQSGTVTVAQSGSDD603 pKa = 2.97QWHH606 pKa = 5.27RR607 pKa = 11.84VVFDD611 pKa = 3.32TTISDD616 pKa = 3.66AVVVMGPISWNGLQAATTRR635 pKa = 11.84VRR637 pKa = 11.84NVTDD641 pKa = 3.26EE642 pKa = 4.15GFEE645 pKa = 3.95FQIDD649 pKa = 3.22EE650 pKa = 4.19WNYY653 pKa = 10.8LDD655 pKa = 4.96GAHH658 pKa = 5.17TTEE661 pKa = 4.54TIGWLAMSEE670 pKa = 4.41GSHH673 pKa = 5.5TLSNGQTIVAGTQIVGTEE691 pKa = 4.19FSSVSFGEE699 pKa = 4.08VLGDD703 pKa = 3.38AVVLAEE709 pKa = 4.21VSTANGKK716 pKa = 9.51DD717 pKa = 3.19AVMTRR722 pKa = 11.84LNDD725 pKa = 3.27VTASGFEE732 pKa = 4.01VQLQEE737 pKa = 4.2EE738 pKa = 4.69EE739 pKa = 4.3ALGAHH744 pKa = 5.92TDD746 pKa = 3.7EE747 pKa = 4.48TVSWIALEE755 pKa = 4.14TGVGDD760 pKa = 3.69GLDD763 pKa = 3.44VFRR766 pKa = 11.84TADD769 pKa = 3.58QLDD772 pKa = 3.98HH773 pKa = 6.83EE774 pKa = 4.8VDD776 pKa = 3.27SFQFNTSFAKK786 pKa = 10.73APVILGDD793 pKa = 3.94LQSKK797 pKa = 10.21DD798 pKa = 3.38GRR800 pKa = 11.84DD801 pKa = 3.44PATLRR806 pKa = 11.84LSQVDD811 pKa = 3.5RR812 pKa = 11.84SGVSLFVEE820 pKa = 4.93EE821 pKa = 4.21EE822 pKa = 4.03TSADD826 pKa = 3.4SEE828 pKa = 4.72VIHH831 pKa = 6.12TEE833 pKa = 3.58EE834 pKa = 3.79TAGYY838 pKa = 8.09VALVEE843 pKa = 4.13GLIFEE848 pKa = 5.4DD849 pKa = 4.13SFLFF853 pKa = 4.46
MM1 pKa = 7.48SNIDD5 pKa = 3.97PSQSSQNPEE14 pKa = 3.88TNQDD18 pKa = 3.82DD19 pKa = 4.46LSLHH23 pKa = 7.0LSTCTCAACMADD35 pKa = 6.09DD36 pKa = 4.95RR37 pKa = 11.84FQDD40 pKa = 3.58DD41 pKa = 3.98VEE43 pKa = 4.69RR44 pKa = 11.84SGNPTSGGGTAAAATGKK61 pKa = 7.26TLRR64 pKa = 11.84EE65 pKa = 3.75MADD68 pKa = 3.87FLEE71 pKa = 4.22MGYY74 pKa = 9.48WGNEE78 pKa = 3.16VGLRR82 pKa = 11.84HH83 pKa = 6.02NVGTTGLDD91 pKa = 3.41PNNGVLHH98 pKa = 5.88YY99 pKa = 9.47NVSGFGPLTYY109 pKa = 10.63GGGSDD114 pKa = 3.86ANGVSSARR122 pKa = 11.84AALIRR127 pKa = 11.84DD128 pKa = 3.45AFDD131 pKa = 2.94VYY133 pKa = 10.9EE134 pKa = 4.47AVLGIQFVEE143 pKa = 4.6TTSTNDD149 pKa = 3.14SVVDD153 pKa = 4.84FFFSDD158 pKa = 3.5NQSGAFAGSTRR169 pKa = 11.84YY170 pKa = 10.38GDD172 pKa = 3.26GTIYY176 pKa = 10.68YY177 pKa = 9.89SYY179 pKa = 11.3INIAASWSGSSSTYY193 pKa = 11.06DD194 pKa = 3.7DD195 pKa = 3.89YY196 pKa = 11.86TLQTIFHH203 pKa = 7.27EE204 pKa = 4.72IGHH207 pKa = 6.51ALGLGHH213 pKa = 6.5QGPYY217 pKa = 10.01NGSASYY223 pKa = 11.6ANDD226 pKa = 4.42AIFALDD232 pKa = 3.13SWQASMMSYY241 pKa = 10.57FSQTEE246 pKa = 3.62NSAISASYY254 pKa = 10.81EE255 pKa = 3.91FLQTPMAVDD264 pKa = 3.78WLALDD269 pKa = 4.84SIYY272 pKa = 10.8GQQGYY277 pKa = 9.76GISNAFVGDD286 pKa = 3.46TVYY289 pKa = 11.14GFNTNIAASEE299 pKa = 3.96SRR301 pKa = 11.84IWNEE305 pKa = 3.12FSAYY309 pKa = 10.06AGRR312 pKa = 11.84TASTIVDD319 pKa = 3.39GGGIDD324 pKa = 3.45TLDD327 pKa = 3.51FSGYY331 pKa = 9.78SANQKK336 pKa = 9.74IDD338 pKa = 3.67LTVQTADD345 pKa = 3.13QTSQNTSNVGGRR357 pKa = 11.84TGNLTLAIGTVIEE370 pKa = 4.1NAIGGSGNDD379 pKa = 3.24YY380 pKa = 10.94FIGNEE385 pKa = 3.63ADD387 pKa = 3.25NVFRR391 pKa = 11.84GGAGNDD397 pKa = 3.16RR398 pKa = 11.84FIGQLGDD405 pKa = 3.63DD406 pKa = 4.17TFHH409 pKa = 7.0GNAGLDD415 pKa = 3.39TVIYY419 pKa = 9.92NLVFGAYY426 pKa = 9.16SFSVLSDD433 pKa = 3.53VIQVIGEE440 pKa = 4.56GIDD443 pKa = 3.57LVYY446 pKa = 10.06DD447 pKa = 4.17TIEE450 pKa = 3.98TFQFSDD456 pKa = 2.78ITYY459 pKa = 9.64SFTDD463 pKa = 3.28IFALVGNAVPTARR476 pKa = 11.84DD477 pKa = 3.23DD478 pKa = 3.74SATVEE483 pKa = 4.12EE484 pKa = 5.26GATITLSVLDD494 pKa = 4.91NDD496 pKa = 4.55TDD498 pKa = 4.31PDD500 pKa = 4.39GDD502 pKa = 4.1PLTITEE508 pKa = 4.54VNGQAIGVNEE518 pKa = 4.25TVDD521 pKa = 3.96LASGASVTLRR531 pKa = 11.84DD532 pKa = 5.0DD533 pKa = 3.57GTLSYY538 pKa = 10.5AQNGVFDD545 pKa = 4.09TLNTGEE551 pKa = 4.34SGGDD555 pKa = 3.21SFEE558 pKa = 4.41YY559 pKa = 10.53AVSDD563 pKa = 3.99GKK565 pKa = 11.26GGAATATVVMTVEE578 pKa = 4.45GAGDD582 pKa = 4.21DD583 pKa = 4.11SPSAAIGQSGTVTVAQSGSDD603 pKa = 2.97QWHH606 pKa = 5.27RR607 pKa = 11.84VVFDD611 pKa = 3.32TTISDD616 pKa = 3.66AVVVMGPISWNGLQAATTRR635 pKa = 11.84VRR637 pKa = 11.84NVTDD641 pKa = 3.26EE642 pKa = 4.15GFEE645 pKa = 3.95FQIDD649 pKa = 3.22EE650 pKa = 4.19WNYY653 pKa = 10.8LDD655 pKa = 4.96GAHH658 pKa = 5.17TTEE661 pKa = 4.54TIGWLAMSEE670 pKa = 4.41GSHH673 pKa = 5.5TLSNGQTIVAGTQIVGTEE691 pKa = 4.19FSSVSFGEE699 pKa = 4.08VLGDD703 pKa = 3.38AVVLAEE709 pKa = 4.21VSTANGKK716 pKa = 9.51DD717 pKa = 3.19AVMTRR722 pKa = 11.84LNDD725 pKa = 3.27VTASGFEE732 pKa = 4.01VQLQEE737 pKa = 4.2EE738 pKa = 4.69EE739 pKa = 4.3ALGAHH744 pKa = 5.92TDD746 pKa = 3.7EE747 pKa = 4.48TVSWIALEE755 pKa = 4.14TGVGDD760 pKa = 3.69GLDD763 pKa = 3.44VFRR766 pKa = 11.84TADD769 pKa = 3.58QLDD772 pKa = 3.98HH773 pKa = 6.83EE774 pKa = 4.8VDD776 pKa = 3.27SFQFNTSFAKK786 pKa = 10.73APVILGDD793 pKa = 3.94LQSKK797 pKa = 10.21DD798 pKa = 3.38GRR800 pKa = 11.84DD801 pKa = 3.44PATLRR806 pKa = 11.84LSQVDD811 pKa = 3.5RR812 pKa = 11.84SGVSLFVEE820 pKa = 4.93EE821 pKa = 4.21EE822 pKa = 4.03TSADD826 pKa = 3.4SEE828 pKa = 4.72VIHH831 pKa = 6.12TEE833 pKa = 3.58EE834 pKa = 3.79TAGYY838 pKa = 8.09VALVEE843 pKa = 4.13GLIFEE848 pKa = 5.4DD849 pKa = 4.13SFLFF853 pKa = 4.46
Molecular weight: 90.05 kDa
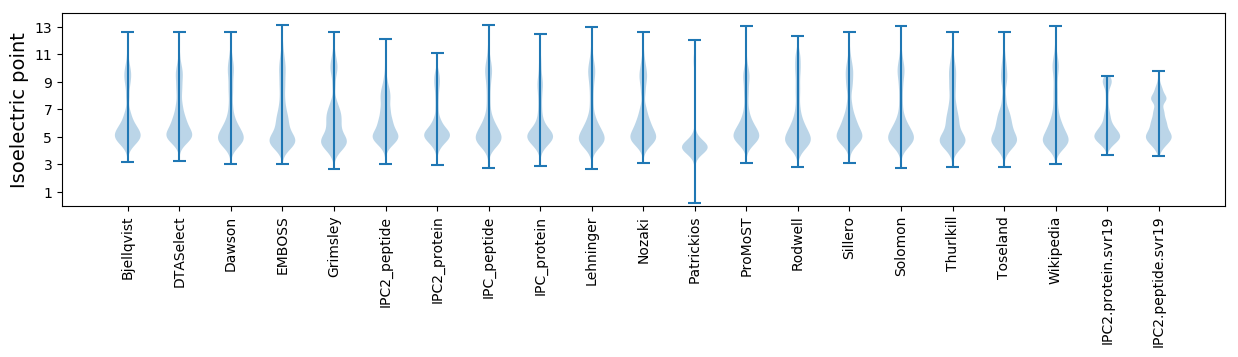
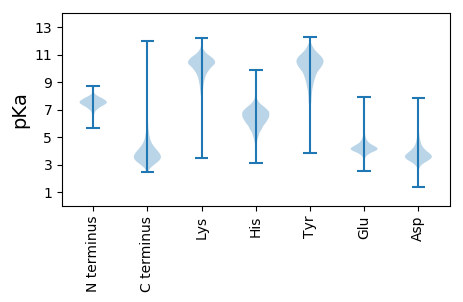
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A640VM94|A0A640VM94_9RHOB Uncharacterized protein OS=Roseobacter cerasinus OX=2602289 GN=So717_02830 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330952 |
39 |
3496 |
306.5 |
33.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.165 ± 0.051 | 0.927 ± 0.012 |
6.201 ± 0.044 | 5.778 ± 0.038 |
3.813 ± 0.022 | 8.518 ± 0.043 |
2.083 ± 0.021 | 5.189 ± 0.026 |
3.056 ± 0.031 | 10.032 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.79 ± 0.02 | 2.579 ± 0.024 |
5.015 ± 0.029 | 3.499 ± 0.02 |
6.478 ± 0.038 | 5.268 ± 0.025 |
5.682 ± 0.034 | 7.319 ± 0.028 |
1.38 ± 0.016 | 2.229 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
