
Mycoplasma iowae DK-CPA
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma iowae
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
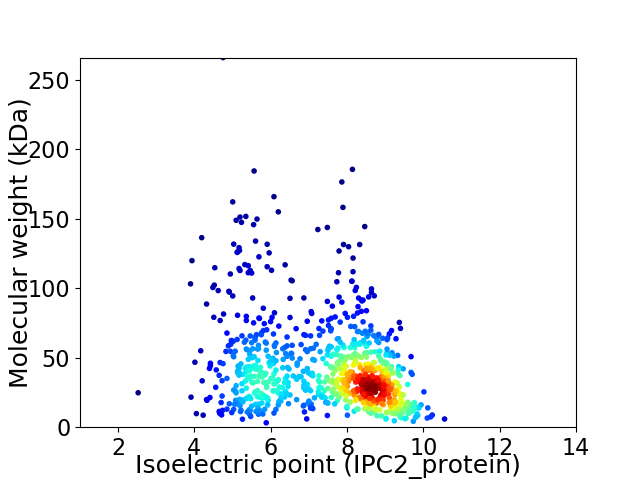
Virtual 2D-PAGE plot for 854 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084U478|A0A084U478_MYCIO Uncharacterized protein OS=Mycoplasma iowae DK-CPA OX=1394179 GN=P271_623 PE=4 SV=1
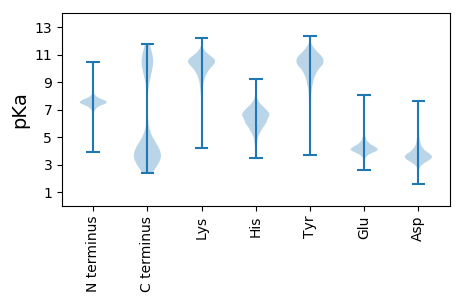
MM1 pKa = 7.55EE2 pKa = 5.55LFEE5 pKa = 4.85KK6 pKa = 10.68LRR8 pKa = 11.84LIIKK12 pKa = 9.2NSKK15 pKa = 9.4FISIISIIYY24 pKa = 9.77IIMLALYY31 pKa = 10.1SVIIIINNQLRR42 pKa = 11.84SQNVHH47 pKa = 4.33VTTDD51 pKa = 3.45GEE53 pKa = 4.34IVINGPVQTLPVGIILFAQICLTIAITLIVLSSTLSINVFMKK95 pKa = 10.55LKK97 pKa = 10.2RR98 pKa = 11.84LSKK101 pKa = 10.69DD102 pKa = 3.22SVHH105 pKa = 6.07GQEE108 pKa = 5.26IYY110 pKa = 11.14DD111 pKa = 3.76NLIICSFLSIIFFGFIFSFILWIKK135 pKa = 8.7SVKK138 pKa = 9.62VSKK141 pKa = 10.71NLEE144 pKa = 3.78NYY146 pKa = 9.33YY147 pKa = 10.86DD148 pKa = 3.94YY149 pKa = 11.79VDD151 pKa = 3.29SLEE154 pKa = 4.14FNEE157 pKa = 5.35NEE159 pKa = 4.05VEE161 pKa = 4.29TKK163 pKa = 10.53KK164 pKa = 10.64RR165 pKa = 11.84FSFFKK170 pKa = 10.46RR171 pKa = 11.84FKK173 pKa = 9.91KK174 pKa = 10.67QKK176 pKa = 10.56DD177 pKa = 3.55DD178 pKa = 3.95FSNDD182 pKa = 3.33EE183 pKa = 4.23IDD185 pKa = 4.3DD186 pKa = 4.0NKK188 pKa = 10.67QIEE191 pKa = 4.5DD192 pKa = 3.34WNIRR196 pKa = 11.84DD197 pKa = 4.04DD198 pKa = 4.35DD199 pKa = 4.91YY200 pKa = 12.05YY201 pKa = 11.06MNSDD205 pKa = 3.18SYY207 pKa = 12.09YY208 pKa = 10.26NNQYY212 pKa = 10.13PDD214 pKa = 3.11NNKK217 pKa = 9.21TYY219 pKa = 11.17NSTQQSDD226 pKa = 4.06YY227 pKa = 11.34QEE229 pKa = 5.45SNDD232 pKa = 4.74LYY234 pKa = 11.5DD235 pKa = 5.35EE236 pKa = 5.25IYY238 pKa = 11.02DD239 pKa = 4.97DD240 pKa = 6.2DD241 pKa = 6.85DD242 pKa = 3.61DD243 pKa = 5.16TIYY246 pKa = 10.68HH247 pKa = 6.63YY248 pKa = 11.56DD249 pKa = 3.8NDD251 pKa = 3.72YY252 pKa = 11.65DD253 pKa = 5.46DD254 pKa = 5.36YY255 pKa = 12.02DD256 pKa = 3.86NEE258 pKa = 4.61PYY260 pKa = 10.45HH261 pKa = 7.63DD262 pKa = 5.8DD263 pKa = 3.52GLGYY267 pKa = 10.54YY268 pKa = 9.8QDD270 pKa = 3.55SQYY273 pKa = 11.67GDD275 pKa = 4.19NNDD278 pKa = 2.73HH279 pKa = 7.3RR280 pKa = 11.84YY281 pKa = 9.47
MM1 pKa = 7.55EE2 pKa = 5.55LFEE5 pKa = 4.85KK6 pKa = 10.68LRR8 pKa = 11.84LIIKK12 pKa = 9.2NSKK15 pKa = 9.4FISIISIIYY24 pKa = 9.77IIMLALYY31 pKa = 10.1SVIIIINNQLRR42 pKa = 11.84SQNVHH47 pKa = 4.33VTTDD51 pKa = 3.45GEE53 pKa = 4.34IVINGPVQTLPVGIILFAQICLTIAITLIVLSSTLSINVFMKK95 pKa = 10.55LKK97 pKa = 10.2RR98 pKa = 11.84LSKK101 pKa = 10.69DD102 pKa = 3.22SVHH105 pKa = 6.07GQEE108 pKa = 5.26IYY110 pKa = 11.14DD111 pKa = 3.76NLIICSFLSIIFFGFIFSFILWIKK135 pKa = 8.7SVKK138 pKa = 9.62VSKK141 pKa = 10.71NLEE144 pKa = 3.78NYY146 pKa = 9.33YY147 pKa = 10.86DD148 pKa = 3.94YY149 pKa = 11.79VDD151 pKa = 3.29SLEE154 pKa = 4.14FNEE157 pKa = 5.35NEE159 pKa = 4.05VEE161 pKa = 4.29TKK163 pKa = 10.53KK164 pKa = 10.64RR165 pKa = 11.84FSFFKK170 pKa = 10.46RR171 pKa = 11.84FKK173 pKa = 9.91KK174 pKa = 10.67QKK176 pKa = 10.56DD177 pKa = 3.55DD178 pKa = 3.95FSNDD182 pKa = 3.33EE183 pKa = 4.23IDD185 pKa = 4.3DD186 pKa = 4.0NKK188 pKa = 10.67QIEE191 pKa = 4.5DD192 pKa = 3.34WNIRR196 pKa = 11.84DD197 pKa = 4.04DD198 pKa = 4.35DD199 pKa = 4.91YY200 pKa = 12.05YY201 pKa = 11.06MNSDD205 pKa = 3.18SYY207 pKa = 12.09YY208 pKa = 10.26NNQYY212 pKa = 10.13PDD214 pKa = 3.11NNKK217 pKa = 9.21TYY219 pKa = 11.17NSTQQSDD226 pKa = 4.06YY227 pKa = 11.34QEE229 pKa = 5.45SNDD232 pKa = 4.74LYY234 pKa = 11.5DD235 pKa = 5.35EE236 pKa = 5.25IYY238 pKa = 11.02DD239 pKa = 4.97DD240 pKa = 6.2DD241 pKa = 6.85DD242 pKa = 3.61DD243 pKa = 5.16TIYY246 pKa = 10.68HH247 pKa = 6.63YY248 pKa = 11.56DD249 pKa = 3.8NDD251 pKa = 3.72YY252 pKa = 11.65DD253 pKa = 5.46DD254 pKa = 5.36YY255 pKa = 12.02DD256 pKa = 3.86NEE258 pKa = 4.61PYY260 pKa = 10.45HH261 pKa = 7.63DD262 pKa = 5.8DD263 pKa = 3.52GLGYY267 pKa = 10.54YY268 pKa = 9.8QDD270 pKa = 3.55SQYY273 pKa = 11.67GDD275 pKa = 4.19NNDD278 pKa = 2.73HH279 pKa = 7.3RR280 pKa = 11.84YY281 pKa = 9.47
Molecular weight: 33.37 kDa
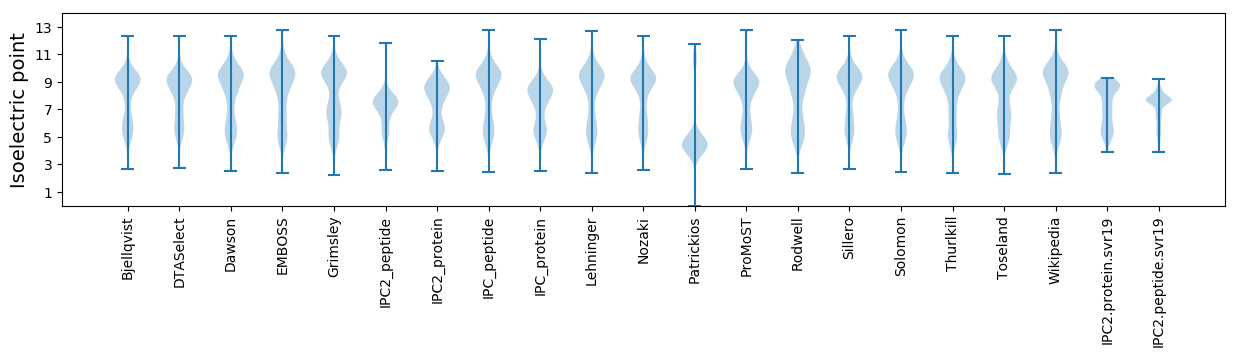
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A084U335|A0A084U335_MYCIO Protein translocase subunit SecY OS=Mycoplasma iowae DK-CPA OX=1394179 GN=secY PE=3 SV=1
MM1 pKa = 7.44ARR3 pKa = 11.84ILGVDD8 pKa = 3.26IPNNKK13 pKa = 9.09RR14 pKa = 11.84VVISLTYY21 pKa = 9.53VYY23 pKa = 10.79GIGRR27 pKa = 11.84SRR29 pKa = 11.84SSEE32 pKa = 3.58ILAKK36 pKa = 10.71AKK38 pKa = 9.82IDD40 pKa = 3.67EE41 pKa = 4.14NRR43 pKa = 11.84KK44 pKa = 9.66VSTLTDD50 pKa = 3.41EE51 pKa = 4.41EE52 pKa = 4.82LATIRR57 pKa = 11.84KK58 pKa = 8.52IAAEE62 pKa = 3.78YY63 pKa = 9.26MIEE66 pKa = 4.21GDD68 pKa = 4.07LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.44TAMNIKK78 pKa = 10.02RR79 pKa = 11.84LMEE82 pKa = 4.29IGCYY86 pKa = 9.42RR87 pKa = 11.84GVRR90 pKa = 11.84HH91 pKa = 6.17RR92 pKa = 11.84RR93 pKa = 11.84SLPVRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.26SNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.46GPRR113 pKa = 11.84KK114 pKa = 7.53TVANKK119 pKa = 10.28KK120 pKa = 9.63IEE122 pKa = 4.32SKK124 pKa = 11.03
MM1 pKa = 7.44ARR3 pKa = 11.84ILGVDD8 pKa = 3.26IPNNKK13 pKa = 9.09RR14 pKa = 11.84VVISLTYY21 pKa = 9.53VYY23 pKa = 10.79GIGRR27 pKa = 11.84SRR29 pKa = 11.84SSEE32 pKa = 3.58ILAKK36 pKa = 10.71AKK38 pKa = 9.82IDD40 pKa = 3.67EE41 pKa = 4.14NRR43 pKa = 11.84KK44 pKa = 9.66VSTLTDD50 pKa = 3.41EE51 pKa = 4.41EE52 pKa = 4.82LATIRR57 pKa = 11.84KK58 pKa = 8.52IAAEE62 pKa = 3.78YY63 pKa = 9.26MIEE66 pKa = 4.21GDD68 pKa = 4.07LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.44TAMNIKK78 pKa = 10.02RR79 pKa = 11.84LMEE82 pKa = 4.29IGCYY86 pKa = 9.42RR87 pKa = 11.84GVRR90 pKa = 11.84HH91 pKa = 6.17RR92 pKa = 11.84RR93 pKa = 11.84SLPVRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.26SNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.46GPRR113 pKa = 11.84KK114 pKa = 7.53TVANKK119 pKa = 10.28KK120 pKa = 9.63IEE122 pKa = 4.32SKK124 pKa = 11.03
Molecular weight: 14.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
325412 |
27 |
2273 |
381.0 |
43.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.14 ± 0.074 | 0.774 ± 0.028 |
5.861 ± 0.064 | 6.114 ± 0.103 |
5.49 ± 0.069 | 4.789 ± 0.088 |
1.149 ± 0.03 | 9.956 ± 0.086 |
10.269 ± 0.097 | 8.827 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.854 ± 0.03 | 8.997 ± 0.109 |
2.6 ± 0.045 | 2.852 ± 0.042 |
2.352 ± 0.046 | 7.399 ± 0.092 |
5.428 ± 0.091 | 5.74 ± 0.062 |
0.939 ± 0.027 | 4.47 ± 0.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
