
Alternaria brassicicola fusarivirus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
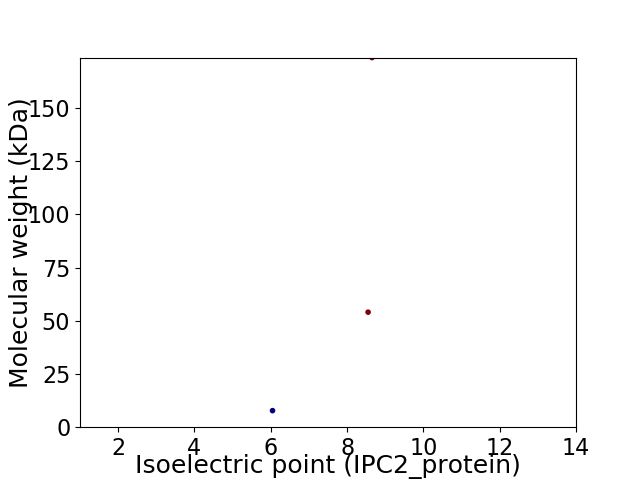
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4D408|A0A0U4D408_9VIRU RNA-directed RNA polymerase OS=Alternaria brassicicola fusarivirus 1 OX=1766767 PE=4 SV=1
MM1 pKa = 7.01MVKK4 pKa = 10.31LVPSSRR10 pKa = 11.84QKK12 pKa = 10.5PEE14 pKa = 3.79KK15 pKa = 10.41EE16 pKa = 3.82EE17 pKa = 4.08LASSRR22 pKa = 11.84QEE24 pKa = 4.1QKK26 pKa = 10.47CLSYY30 pKa = 11.14VLSLSGTGFMSVQGGKK46 pKa = 7.39PQCRR50 pKa = 11.84RR51 pKa = 11.84VEE53 pKa = 4.03TCDD56 pKa = 3.54NSEE59 pKa = 4.66CNDD62 pKa = 3.55CEE64 pKa = 4.56LFVPVSS70 pKa = 3.38
MM1 pKa = 7.01MVKK4 pKa = 10.31LVPSSRR10 pKa = 11.84QKK12 pKa = 10.5PEE14 pKa = 3.79KK15 pKa = 10.41EE16 pKa = 3.82EE17 pKa = 4.08LASSRR22 pKa = 11.84QEE24 pKa = 4.1QKK26 pKa = 10.47CLSYY30 pKa = 11.14VLSLSGTGFMSVQGGKK46 pKa = 7.39PQCRR50 pKa = 11.84RR51 pKa = 11.84VEE53 pKa = 4.03TCDD56 pKa = 3.54NSEE59 pKa = 4.66CNDD62 pKa = 3.55CEE64 pKa = 4.56LFVPVSS70 pKa = 3.38
Molecular weight: 7.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4B4N9|A0A0U4B4N9_9VIRU Uncharacterized protein OS=Alternaria brassicicola fusarivirus 1 OX=1766767 PE=4 SV=1
MM1 pKa = 7.32SANKK5 pKa = 9.68IFEE8 pKa = 4.17GKK10 pKa = 10.18RR11 pKa = 11.84IVKK14 pKa = 9.97ARR16 pKa = 11.84EE17 pKa = 3.52QLASVEE23 pKa = 4.04LTAQQVKK30 pKa = 10.47SSTVWLVRR38 pKa = 11.84EE39 pKa = 4.31PGWIAGEE46 pKa = 4.08EE47 pKa = 4.1KK48 pKa = 10.97LNGWFLEE55 pKa = 4.25IQGSDD60 pKa = 2.43IVLCVTTLFKK70 pKa = 11.58GEE72 pKa = 4.18VTSEE76 pKa = 3.5RR77 pKa = 11.84WFPPSPVFKK86 pKa = 10.0WKK88 pKa = 10.39KK89 pKa = 9.02EE90 pKa = 3.85YY91 pKa = 10.14QSRR94 pKa = 11.84DD95 pKa = 3.16KK96 pKa = 11.09EE97 pKa = 4.51VASEE101 pKa = 3.8EE102 pKa = 4.33AYY104 pKa = 10.11KK105 pKa = 10.64KK106 pKa = 10.84VIVARR111 pKa = 11.84MADD114 pKa = 3.87EE115 pKa = 4.52NLLGYY120 pKa = 10.93YY121 pKa = 10.95DD122 pKa = 4.64MLTPATPLVNRR133 pKa = 11.84LSASFKK139 pKa = 8.0WWKK142 pKa = 8.96TGGNAPIIPFSPTNSKK158 pKa = 10.82SEE160 pKa = 3.82IVGRR164 pKa = 11.84IAHH167 pKa = 6.61FEE169 pKa = 3.99TAFAALNNNPPFALDD184 pKa = 3.18GLLALRR190 pKa = 11.84VLKK193 pKa = 10.71DD194 pKa = 3.27QLPNMKK200 pKa = 10.27DD201 pKa = 2.78RR202 pKa = 11.84GAQNVSRR209 pKa = 11.84FLTTLVKK216 pKa = 10.09AWAMSKK222 pKa = 10.14QPSHH226 pKa = 7.01FKK228 pKa = 11.08GPLSKK233 pKa = 9.94ISYY236 pKa = 7.0PTEE239 pKa = 4.27KK240 pKa = 10.35EE241 pKa = 3.8GLPADD246 pKa = 4.03RR247 pKa = 11.84AKK249 pKa = 10.49IFHH252 pKa = 6.46NLICGKK258 pKa = 10.15GDD260 pKa = 3.8EE261 pKa = 4.37ITRR264 pKa = 11.84GLGFCLWAPPRR275 pKa = 11.84ITVTTDD281 pKa = 2.73AEE283 pKa = 4.79GFTSKK288 pKa = 9.24QRR290 pKa = 11.84KK291 pKa = 7.02KK292 pKa = 10.2HH293 pKa = 4.67EE294 pKa = 4.32EE295 pKa = 3.88KK296 pKa = 10.76KK297 pKa = 10.66IMLDD301 pKa = 3.19IGPDD305 pKa = 3.28IKK307 pKa = 11.18VGAALDD313 pKa = 3.9AEE315 pKa = 4.25PLEE318 pKa = 4.6GSFAEE323 pKa = 4.66SSAAGASRR331 pKa = 11.84SHH333 pKa = 5.91MKK335 pKa = 10.52KK336 pKa = 10.5NSDD339 pKa = 3.49PEE341 pKa = 4.33ANVLKK346 pKa = 10.66EE347 pKa = 4.36SGPKK351 pKa = 8.88PDD353 pKa = 3.63NVQEE357 pKa = 4.17EE358 pKa = 4.98VVDD361 pKa = 5.28EE362 pKa = 4.34AADD365 pKa = 3.44WLKK368 pKa = 11.44NSFTMVNFRR377 pKa = 11.84RR378 pKa = 11.84FYY380 pKa = 10.36RR381 pKa = 11.84RR382 pKa = 11.84VKK384 pKa = 10.63RR385 pKa = 11.84SVTTIKK391 pKa = 10.68EE392 pKa = 4.17SVTYY396 pKa = 10.04EE397 pKa = 3.7GVGINRR403 pKa = 11.84PFMEE407 pKa = 4.43KK408 pKa = 10.55AKK410 pKa = 8.77NTVVFTVTSAVKK422 pKa = 10.28ACATVPIAAAMGVWYY437 pKa = 9.82GVKK440 pKa = 10.27EE441 pKa = 4.04PFVSIKK447 pKa = 10.64DD448 pKa = 3.64FPTPSGKK455 pKa = 9.77VEE457 pKa = 4.15MAKK460 pKa = 10.2HH461 pKa = 5.11VADD464 pKa = 3.75CAVYY468 pKa = 10.33SVTSVVTSTISGIFAGLASATRR490 pKa = 3.68
MM1 pKa = 7.32SANKK5 pKa = 9.68IFEE8 pKa = 4.17GKK10 pKa = 10.18RR11 pKa = 11.84IVKK14 pKa = 9.97ARR16 pKa = 11.84EE17 pKa = 3.52QLASVEE23 pKa = 4.04LTAQQVKK30 pKa = 10.47SSTVWLVRR38 pKa = 11.84EE39 pKa = 4.31PGWIAGEE46 pKa = 4.08EE47 pKa = 4.1KK48 pKa = 10.97LNGWFLEE55 pKa = 4.25IQGSDD60 pKa = 2.43IVLCVTTLFKK70 pKa = 11.58GEE72 pKa = 4.18VTSEE76 pKa = 3.5RR77 pKa = 11.84WFPPSPVFKK86 pKa = 10.0WKK88 pKa = 10.39KK89 pKa = 9.02EE90 pKa = 3.85YY91 pKa = 10.14QSRR94 pKa = 11.84DD95 pKa = 3.16KK96 pKa = 11.09EE97 pKa = 4.51VASEE101 pKa = 3.8EE102 pKa = 4.33AYY104 pKa = 10.11KK105 pKa = 10.64KK106 pKa = 10.84VIVARR111 pKa = 11.84MADD114 pKa = 3.87EE115 pKa = 4.52NLLGYY120 pKa = 10.93YY121 pKa = 10.95DD122 pKa = 4.64MLTPATPLVNRR133 pKa = 11.84LSASFKK139 pKa = 8.0WWKK142 pKa = 8.96TGGNAPIIPFSPTNSKK158 pKa = 10.82SEE160 pKa = 3.82IVGRR164 pKa = 11.84IAHH167 pKa = 6.61FEE169 pKa = 3.99TAFAALNNNPPFALDD184 pKa = 3.18GLLALRR190 pKa = 11.84VLKK193 pKa = 10.71DD194 pKa = 3.27QLPNMKK200 pKa = 10.27DD201 pKa = 2.78RR202 pKa = 11.84GAQNVSRR209 pKa = 11.84FLTTLVKK216 pKa = 10.09AWAMSKK222 pKa = 10.14QPSHH226 pKa = 7.01FKK228 pKa = 11.08GPLSKK233 pKa = 9.94ISYY236 pKa = 7.0PTEE239 pKa = 4.27KK240 pKa = 10.35EE241 pKa = 3.8GLPADD246 pKa = 4.03RR247 pKa = 11.84AKK249 pKa = 10.49IFHH252 pKa = 6.46NLICGKK258 pKa = 10.15GDD260 pKa = 3.8EE261 pKa = 4.37ITRR264 pKa = 11.84GLGFCLWAPPRR275 pKa = 11.84ITVTTDD281 pKa = 2.73AEE283 pKa = 4.79GFTSKK288 pKa = 9.24QRR290 pKa = 11.84KK291 pKa = 7.02KK292 pKa = 10.2HH293 pKa = 4.67EE294 pKa = 4.32EE295 pKa = 3.88KK296 pKa = 10.76KK297 pKa = 10.66IMLDD301 pKa = 3.19IGPDD305 pKa = 3.28IKK307 pKa = 11.18VGAALDD313 pKa = 3.9AEE315 pKa = 4.25PLEE318 pKa = 4.6GSFAEE323 pKa = 4.66SSAAGASRR331 pKa = 11.84SHH333 pKa = 5.91MKK335 pKa = 10.52KK336 pKa = 10.5NSDD339 pKa = 3.49PEE341 pKa = 4.33ANVLKK346 pKa = 10.66EE347 pKa = 4.36SGPKK351 pKa = 8.88PDD353 pKa = 3.63NVQEE357 pKa = 4.17EE358 pKa = 4.98VVDD361 pKa = 5.28EE362 pKa = 4.34AADD365 pKa = 3.44WLKK368 pKa = 11.44NSFTMVNFRR377 pKa = 11.84RR378 pKa = 11.84FYY380 pKa = 10.36RR381 pKa = 11.84RR382 pKa = 11.84VKK384 pKa = 10.63RR385 pKa = 11.84SVTTIKK391 pKa = 10.68EE392 pKa = 4.17SVTYY396 pKa = 10.04EE397 pKa = 3.7GVGINRR403 pKa = 11.84PFMEE407 pKa = 4.43KK408 pKa = 10.55AKK410 pKa = 8.77NTVVFTVTSAVKK422 pKa = 10.28ACATVPIAAAMGVWYY437 pKa = 9.82GVKK440 pKa = 10.27EE441 pKa = 4.04PFVSIKK447 pKa = 10.64DD448 pKa = 3.64FPTPSGKK455 pKa = 9.77VEE457 pKa = 4.15MAKK460 pKa = 10.2HH461 pKa = 5.11VADD464 pKa = 3.75CAVYY468 pKa = 10.33SVTSVVTSTISGIFAGLASATRR490 pKa = 3.68
Molecular weight: 54.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2082 |
70 |
1522 |
694.0 |
78.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.916 ± 1.404 | 0.865 ± 1.023 |
4.803 ± 0.606 | 6.148 ± 0.874 |
5.091 ± 0.392 | 5.812 ± 0.241 |
1.873 ± 0.486 | 5.331 ± 0.928 |
7.301 ± 0.793 | 9.75 ± 1.597 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.594 ± 0.274 | 3.65 ± 0.141 |
4.947 ± 0.417 | 2.257 ± 0.755 |
5.764 ± 0.512 | 7.349 ± 1.173 |
6.34 ± 0.556 | 7.973 ± 0.487 |
2.354 ± 0.392 | 2.882 ± 0.605 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
