
Methylobacterium sp. YIM 132548
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium; unclassified Methylobacterium
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
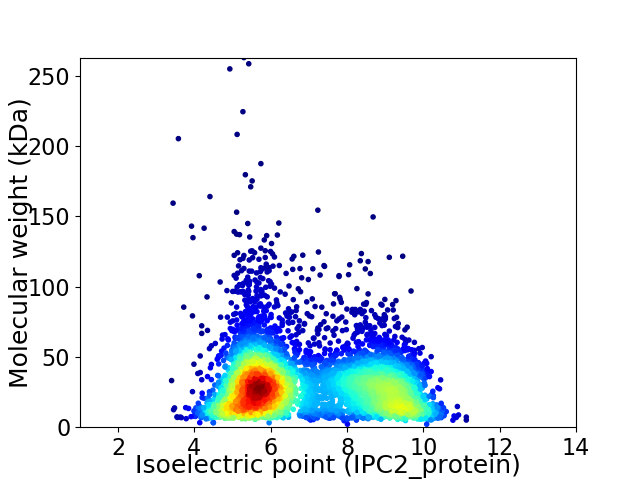
Virtual 2D-PAGE plot for 5304 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6MJM9|A0A6N6MJM9_9RHIZ Acyl-CoA dehydrogenase OS=Methylobacterium sp. YIM 132548 OX=2615211 GN=F6X51_23340 PE=3 SV=1
AA1 pKa = 7.63FSVTPGTALGDD12 pKa = 3.48GPYY15 pKa = 10.88AFTATATDD23 pKa = 3.35AAGNLGAPSAPVSLTIDD40 pKa = 3.61TAVPAAPVVTSAGGATSDD58 pKa = 3.48TTPAITGTGEE68 pKa = 3.74AGATVTLLDD77 pKa = 3.34GTAPVGTATVGQNGGFTVSPGAPLGEE103 pKa = 4.69GPHH106 pKa = 5.93NLSVQLTDD114 pKa = 3.14AAGNVGAPSPAVAVTIDD131 pKa = 3.75SLAPTIPVVTGGGGATADD149 pKa = 3.52TTPAVTGTAEE159 pKa = 4.17PGATVTLLNGTTALGTATVNPDD181 pKa = 2.55GSFTVSPTTPLGQGTYY197 pKa = 9.87DD198 pKa = 3.46LSVRR202 pKa = 11.84VTDD205 pKa = 3.87AAGNTSPVTAAIGVTVDD222 pKa = 3.23TTLPTTPTLTAPAGPTSDD240 pKa = 3.61STPTLTGTAEE250 pKa = 4.13ANSTVTIANGNTVLGTVTADD270 pKa = 2.85SGGAFSFTPVPAFGDD285 pKa = 3.4GTYY288 pKa = 11.45ALTATVRR295 pKa = 11.84DD296 pKa = 3.39AAGNVGAPSAPVSLTIDD313 pKa = 3.35TAAPVMPVLTSAGGPTSSAAPAITGTGEE341 pKa = 3.06IGARR345 pKa = 11.84VTLLDD350 pKa = 3.17GTAPVGTAIVDD361 pKa = 3.42QDD363 pKa = 3.44GSFTVSPGAPLGEE376 pKa = 4.69GPHH379 pKa = 5.93NLSVQLTDD387 pKa = 3.14AAGNVGAPSMAITVTVDD404 pKa = 2.78VTAPAAPVFTTASGVTADD422 pKa = 4.31ATPAIAGTAEE432 pKa = 3.99AGATVTVFDD441 pKa = 5.57DD442 pKa = 4.03GTPLGTVQANARR454 pKa = 11.84GAWSFTPGTALANGPNLITATATDD478 pKa = 3.72AVGNISPAATGPTLTVDD495 pKa = 3.91TGAPPAPVLTIAPGPTNDD513 pKa = 3.54NTPTITGTAEE523 pKa = 3.8ANSTVTITSGGILIATVTADD543 pKa = 2.89AGGAFSVTPGTALGDD558 pKa = 3.48GPYY561 pKa = 10.88AFTATATDD569 pKa = 3.35AAGNLGAPSAPVSLTIDD586 pKa = 3.61TAVPAAPVVTSAGGATSDD604 pKa = 3.48TTPAITGTGEE614 pKa = 3.74AGATVTLLDD623 pKa = 3.34GTAPVGTATVGQNGGFTVSPGAPLGEE649 pKa = 4.69GPHH652 pKa = 6.09NLSVHH657 pKa = 5.55LTDD660 pKa = 4.04AAGNVGAPSPAVAVTVDD677 pKa = 3.68TNVDD681 pKa = 3.65VPPALAFTVATTADD695 pKa = 3.17GRR697 pKa = 11.84LNATEE702 pKa = 4.13AQATAFTIAGLDD714 pKa = 3.29AGTTGTATFTDD725 pKa = 3.8ASTASVSVAIGANGTYY741 pKa = 10.52AADD744 pKa = 3.94LSGLDD749 pKa = 3.84GPVTTSLRR757 pKa = 11.84VTDD760 pKa = 3.9PAGNAGTIAGPSLTLDD776 pKa = 3.19TLAPVGTAVADD787 pKa = 3.77ASGGPEE793 pKa = 3.87VASFTYY799 pKa = 9.52TVSFPEE805 pKa = 3.88AVANVGAEE813 pKa = 4.07DD814 pKa = 3.84FTLTGTNGATGTIAGVTGSGGTYY837 pKa = 8.04TVTVAEE843 pKa = 4.23VSGAGTLSLGLAAGSDD859 pKa = 3.54ITDD862 pKa = 3.85LAGNAASLTPADD874 pKa = 3.75RR875 pKa = 11.84AVIGVAPVLPVITGYY890 pKa = 9.3TIDD893 pKa = 3.91TGVPSDD899 pKa = 5.27GITGDD904 pKa = 3.63PTPTLTGIGAAGSTVTVTYY923 pKa = 8.86ATAAGPAQATGTVLADD939 pKa = 3.89GTWSLTLPTLADD951 pKa = 3.29GDD953 pKa = 4.36YY954 pKa = 11.33SFTASLATAGGTPIGTSAPLALTIDD979 pKa = 3.83TGVDD983 pKa = 2.69AGTPTTLVVDD993 pKa = 4.55GTADD997 pKa = 3.55GLINAAEE1004 pKa = 4.18ATAVAYY1010 pKa = 8.09TVAGLDD1016 pKa = 3.33AGTSGTITFRR1026 pKa = 11.84DD1027 pKa = 5.03GIRR1030 pKa = 11.84ATEE1033 pKa = 4.08VAVSQNGTFAVDD1045 pKa = 3.49LSGFSGPVSANLALRR1060 pKa = 11.84DD1061 pKa = 3.8VAGNAATLAGNTVSLATGAPGAPVITGLADD1091 pKa = 3.39DD1092 pKa = 4.78TGTLGDD1098 pKa = 4.4TLTADD1103 pKa = 3.88TNPTLSGLADD1113 pKa = 4.21PGSAIRR1119 pKa = 11.84LDD1121 pKa = 3.71FTTAAGPQFVEE1132 pKa = 5.16TVAGASGAWTATLPLLPDD1150 pKa = 3.74GAYY1153 pKa = 10.38AVTASARR1160 pKa = 11.84DD1161 pKa = 3.43AAGNASVASAPFPISIDD1178 pKa = 3.08ATAPAGTAGADD1189 pKa = 3.46TAGGLTAEE1197 pKa = 4.45TFTYY1201 pKa = 9.7TVSFPEE1207 pKa = 3.88AVANVGAEE1215 pKa = 4.16DD1216 pKa = 4.6FSLTGTNGATGTIAGVTGSGGTYY1239 pKa = 7.89TVTVAGVSGAGTLSLGLAAGSDD1261 pKa = 3.23IADD1264 pKa = 3.9RR1265 pKa = 11.84AGNLASLTPATRR1277 pKa = 11.84NVAVGPDD1284 pKa = 3.32ATVPTTISGFTEE1296 pKa = 4.37DD1297 pKa = 4.53SGVAGDD1303 pKa = 5.62GITSDD1308 pKa = 3.75ATPTLTGTGNAGGTVTVTFATAAGAQTLTGPVDD1341 pKa = 3.95ALGNWTVPVPEE1352 pKa = 4.87LADD1355 pKa = 3.31GRR1357 pKa = 11.84YY1358 pKa = 10.03SFTATTSGPSGAPGGTSAPLALTIDD1383 pKa = 4.12TVADD1387 pKa = 3.84ALPAVALALDD1397 pKa = 4.68GSADD1401 pKa = 3.68GVLTPAEE1408 pKa = 4.0AATARR1413 pKa = 11.84FTLSGLDD1420 pKa = 3.52AGSGAAVTFTDD1431 pKa = 3.67AAGTRR1436 pKa = 11.84VAATASADD1444 pKa = 3.25GTYY1447 pKa = 10.03TVDD1450 pKa = 3.63LSGLIGPVTSSVVVTDD1466 pKa = 3.74AAGNSAVGSGNPIVIDD1482 pKa = 4.02AGTTTPPRR1490 pKa = 11.84TPVPTITGIQEE1501 pKa = 4.03DD1502 pKa = 4.46TGTPGDD1508 pKa = 5.07GITSDD1513 pKa = 3.77ASPILVGTASPGSTVTVSYY1532 pKa = 10.14TGAAGGAATATGTVGTDD1549 pKa = 3.71GAWQIALPALADD1561 pKa = 3.69GSYY1564 pKa = 11.01SFVATALAPGSTSSPASQALPVVIDD1589 pKa = 3.76TVVPAAPVFGGIVGDD1604 pKa = 4.6DD1605 pKa = 3.59GDD1607 pKa = 4.75GVVTDD1612 pKa = 5.13ASPTLTGTAEE1622 pKa = 4.31PGSTVTVTYY1631 pKa = 7.42EE1632 pKa = 4.19TPQGPGSATGTTGTDD1647 pKa = 3.37GRR1649 pKa = 11.84WTVDD1653 pKa = 3.06IPTLPDD1659 pKa = 3.11GSYY1662 pKa = 10.83SLTVGTTDD1670 pKa = 3.18AAGNAGAPAAPVSLTINTGTGGGGTGGGGTGGGGTGGGSTGGGGTGGDD1718 pKa = 3.8GGTGGSGGSGGSGGTAPVLVGDD1740 pKa = 4.64LNQTLAHH1747 pKa = 6.46APAVSGNLLANDD1759 pKa = 3.74TGAALKK1765 pKa = 8.96VTAVQFSGGLPVAVPTEE1782 pKa = 4.13GTVKK1786 pKa = 10.27VVSDD1790 pKa = 4.02HH1791 pKa = 5.83GTLTVSADD1799 pKa = 3.06GSYY1802 pKa = 10.53SYY1804 pKa = 10.89QAIGANNLDD1813 pKa = 3.67DD1814 pKa = 3.9TVRR1817 pKa = 11.84TAEE1820 pKa = 4.09HH1821 pKa = 5.4FTYY1824 pKa = 10.23TVTDD1828 pKa = 3.28AAGAVGQSSLDD1839 pKa = 3.3ISLGGQAPQASASFGFAFTEE1859 pKa = 4.23ARR1861 pKa = 11.84VTLAGEE1867 pKa = 4.14ALVLVGPDD1875 pKa = 3.04GVTRR1879 pKa = 11.84DD1880 pKa = 3.08ISGIDD1885 pKa = 3.24TLRR1888 pKa = 11.84FTDD1891 pKa = 5.98GEE1893 pKa = 4.1IQNNDD1898 pKa = 2.76GHH1900 pKa = 7.09HH1901 pKa = 5.99VVDD1904 pKa = 5.03DD1905 pKa = 3.66VFYY1908 pKa = 10.62YY1909 pKa = 11.16ANNLDD1914 pKa = 3.19VWRR1917 pKa = 11.84AHH1919 pKa = 6.07VDD1921 pKa = 2.97ADD1923 pKa = 3.56THH1925 pKa = 6.61YY1926 pKa = 9.77EE1927 pKa = 3.98TFGWKK1932 pKa = 9.75EE1933 pKa = 3.67GRR1935 pKa = 11.84DD1936 pKa = 3.63PNAYY1940 pKa = 9.38FHH1942 pKa = 6.25TRR1944 pKa = 11.84AYY1946 pKa = 10.45LAEE1949 pKa = 4.43NPDD1952 pKa = 3.21VAAANINPLEE1962 pKa = 4.59HH1963 pKa = 6.65YY1964 pKa = 8.81LTFGEE1969 pKa = 4.35RR1970 pKa = 11.84EE1971 pKa = 4.18GRR1973 pKa = 11.84SPSPGFSAEE1982 pKa = 4.42AYY1984 pKa = 9.85LHH1986 pKa = 6.6ANPDD1990 pKa = 3.16VAAAGINALAHH2001 pKa = 6.02YY2002 pKa = 7.44LTNGQDD2008 pKa = 3.03EE2009 pKa = 4.65GRR2011 pKa = 11.84TVFAGEE2017 pKa = 4.12SEE2019 pKa = 4.53GPGRR2023 pKa = 11.84VIGDD2027 pKa = 3.85FDD2029 pKa = 5.77ADD2031 pKa = 4.43FYY2033 pKa = 11.05LAQYY2037 pKa = 10.57ADD2039 pKa = 3.38VAAAVPAGRR2048 pKa = 11.84EE2049 pKa = 3.8AASYY2053 pKa = 10.92ALEE2056 pKa = 4.66HH2057 pKa = 6.48YY2058 pKa = 8.73LTFGARR2064 pKa = 11.84EE2065 pKa = 3.95LRR2067 pKa = 11.84NPNAAFDD2074 pKa = 3.65TAYY2077 pKa = 10.97YY2078 pKa = 9.65LQQNPEE2084 pKa = 4.01VAASGANPLLHH2095 pKa = 5.49YY2096 pKa = 10.76QEE2098 pKa = 5.03VGWHH2102 pKa = 5.97EE2103 pKa = 4.01GRR2105 pKa = 11.84NPSASFNTNAYY2116 pKa = 8.95LASHH2120 pKa = 7.15PEE2122 pKa = 3.86VAQAGIDD2129 pKa = 3.77PLEE2132 pKa = 5.33HH2133 pKa = 6.1YY2134 pKa = 10.11LQTGGHH2140 pKa = 6.66ALAA2143 pKa = 5.38
AA1 pKa = 7.63FSVTPGTALGDD12 pKa = 3.48GPYY15 pKa = 10.88AFTATATDD23 pKa = 3.35AAGNLGAPSAPVSLTIDD40 pKa = 3.61TAVPAAPVVTSAGGATSDD58 pKa = 3.48TTPAITGTGEE68 pKa = 3.74AGATVTLLDD77 pKa = 3.34GTAPVGTATVGQNGGFTVSPGAPLGEE103 pKa = 4.69GPHH106 pKa = 5.93NLSVQLTDD114 pKa = 3.14AAGNVGAPSPAVAVTIDD131 pKa = 3.75SLAPTIPVVTGGGGATADD149 pKa = 3.52TTPAVTGTAEE159 pKa = 4.17PGATVTLLNGTTALGTATVNPDD181 pKa = 2.55GSFTVSPTTPLGQGTYY197 pKa = 9.87DD198 pKa = 3.46LSVRR202 pKa = 11.84VTDD205 pKa = 3.87AAGNTSPVTAAIGVTVDD222 pKa = 3.23TTLPTTPTLTAPAGPTSDD240 pKa = 3.61STPTLTGTAEE250 pKa = 4.13ANSTVTIANGNTVLGTVTADD270 pKa = 2.85SGGAFSFTPVPAFGDD285 pKa = 3.4GTYY288 pKa = 11.45ALTATVRR295 pKa = 11.84DD296 pKa = 3.39AAGNVGAPSAPVSLTIDD313 pKa = 3.35TAAPVMPVLTSAGGPTSSAAPAITGTGEE341 pKa = 3.06IGARR345 pKa = 11.84VTLLDD350 pKa = 3.17GTAPVGTAIVDD361 pKa = 3.42QDD363 pKa = 3.44GSFTVSPGAPLGEE376 pKa = 4.69GPHH379 pKa = 5.93NLSVQLTDD387 pKa = 3.14AAGNVGAPSMAITVTVDD404 pKa = 2.78VTAPAAPVFTTASGVTADD422 pKa = 4.31ATPAIAGTAEE432 pKa = 3.99AGATVTVFDD441 pKa = 5.57DD442 pKa = 4.03GTPLGTVQANARR454 pKa = 11.84GAWSFTPGTALANGPNLITATATDD478 pKa = 3.72AVGNISPAATGPTLTVDD495 pKa = 3.91TGAPPAPVLTIAPGPTNDD513 pKa = 3.54NTPTITGTAEE523 pKa = 3.8ANSTVTITSGGILIATVTADD543 pKa = 2.89AGGAFSVTPGTALGDD558 pKa = 3.48GPYY561 pKa = 10.88AFTATATDD569 pKa = 3.35AAGNLGAPSAPVSLTIDD586 pKa = 3.61TAVPAAPVVTSAGGATSDD604 pKa = 3.48TTPAITGTGEE614 pKa = 3.74AGATVTLLDD623 pKa = 3.34GTAPVGTATVGQNGGFTVSPGAPLGEE649 pKa = 4.69GPHH652 pKa = 6.09NLSVHH657 pKa = 5.55LTDD660 pKa = 4.04AAGNVGAPSPAVAVTVDD677 pKa = 3.68TNVDD681 pKa = 3.65VPPALAFTVATTADD695 pKa = 3.17GRR697 pKa = 11.84LNATEE702 pKa = 4.13AQATAFTIAGLDD714 pKa = 3.29AGTTGTATFTDD725 pKa = 3.8ASTASVSVAIGANGTYY741 pKa = 10.52AADD744 pKa = 3.94LSGLDD749 pKa = 3.84GPVTTSLRR757 pKa = 11.84VTDD760 pKa = 3.9PAGNAGTIAGPSLTLDD776 pKa = 3.19TLAPVGTAVADD787 pKa = 3.77ASGGPEE793 pKa = 3.87VASFTYY799 pKa = 9.52TVSFPEE805 pKa = 3.88AVANVGAEE813 pKa = 4.07DD814 pKa = 3.84FTLTGTNGATGTIAGVTGSGGTYY837 pKa = 8.04TVTVAEE843 pKa = 4.23VSGAGTLSLGLAAGSDD859 pKa = 3.54ITDD862 pKa = 3.85LAGNAASLTPADD874 pKa = 3.75RR875 pKa = 11.84AVIGVAPVLPVITGYY890 pKa = 9.3TIDD893 pKa = 3.91TGVPSDD899 pKa = 5.27GITGDD904 pKa = 3.63PTPTLTGIGAAGSTVTVTYY923 pKa = 8.86ATAAGPAQATGTVLADD939 pKa = 3.89GTWSLTLPTLADD951 pKa = 3.29GDD953 pKa = 4.36YY954 pKa = 11.33SFTASLATAGGTPIGTSAPLALTIDD979 pKa = 3.83TGVDD983 pKa = 2.69AGTPTTLVVDD993 pKa = 4.55GTADD997 pKa = 3.55GLINAAEE1004 pKa = 4.18ATAVAYY1010 pKa = 8.09TVAGLDD1016 pKa = 3.33AGTSGTITFRR1026 pKa = 11.84DD1027 pKa = 5.03GIRR1030 pKa = 11.84ATEE1033 pKa = 4.08VAVSQNGTFAVDD1045 pKa = 3.49LSGFSGPVSANLALRR1060 pKa = 11.84DD1061 pKa = 3.8VAGNAATLAGNTVSLATGAPGAPVITGLADD1091 pKa = 3.39DD1092 pKa = 4.78TGTLGDD1098 pKa = 4.4TLTADD1103 pKa = 3.88TNPTLSGLADD1113 pKa = 4.21PGSAIRR1119 pKa = 11.84LDD1121 pKa = 3.71FTTAAGPQFVEE1132 pKa = 5.16TVAGASGAWTATLPLLPDD1150 pKa = 3.74GAYY1153 pKa = 10.38AVTASARR1160 pKa = 11.84DD1161 pKa = 3.43AAGNASVASAPFPISIDD1178 pKa = 3.08ATAPAGTAGADD1189 pKa = 3.46TAGGLTAEE1197 pKa = 4.45TFTYY1201 pKa = 9.7TVSFPEE1207 pKa = 3.88AVANVGAEE1215 pKa = 4.16DD1216 pKa = 4.6FSLTGTNGATGTIAGVTGSGGTYY1239 pKa = 7.89TVTVAGVSGAGTLSLGLAAGSDD1261 pKa = 3.23IADD1264 pKa = 3.9RR1265 pKa = 11.84AGNLASLTPATRR1277 pKa = 11.84NVAVGPDD1284 pKa = 3.32ATVPTTISGFTEE1296 pKa = 4.37DD1297 pKa = 4.53SGVAGDD1303 pKa = 5.62GITSDD1308 pKa = 3.75ATPTLTGTGNAGGTVTVTFATAAGAQTLTGPVDD1341 pKa = 3.95ALGNWTVPVPEE1352 pKa = 4.87LADD1355 pKa = 3.31GRR1357 pKa = 11.84YY1358 pKa = 10.03SFTATTSGPSGAPGGTSAPLALTIDD1383 pKa = 4.12TVADD1387 pKa = 3.84ALPAVALALDD1397 pKa = 4.68GSADD1401 pKa = 3.68GVLTPAEE1408 pKa = 4.0AATARR1413 pKa = 11.84FTLSGLDD1420 pKa = 3.52AGSGAAVTFTDD1431 pKa = 3.67AAGTRR1436 pKa = 11.84VAATASADD1444 pKa = 3.25GTYY1447 pKa = 10.03TVDD1450 pKa = 3.63LSGLIGPVTSSVVVTDD1466 pKa = 3.74AAGNSAVGSGNPIVIDD1482 pKa = 4.02AGTTTPPRR1490 pKa = 11.84TPVPTITGIQEE1501 pKa = 4.03DD1502 pKa = 4.46TGTPGDD1508 pKa = 5.07GITSDD1513 pKa = 3.77ASPILVGTASPGSTVTVSYY1532 pKa = 10.14TGAAGGAATATGTVGTDD1549 pKa = 3.71GAWQIALPALADD1561 pKa = 3.69GSYY1564 pKa = 11.01SFVATALAPGSTSSPASQALPVVIDD1589 pKa = 3.76TVVPAAPVFGGIVGDD1604 pKa = 4.6DD1605 pKa = 3.59GDD1607 pKa = 4.75GVVTDD1612 pKa = 5.13ASPTLTGTAEE1622 pKa = 4.31PGSTVTVTYY1631 pKa = 7.42EE1632 pKa = 4.19TPQGPGSATGTTGTDD1647 pKa = 3.37GRR1649 pKa = 11.84WTVDD1653 pKa = 3.06IPTLPDD1659 pKa = 3.11GSYY1662 pKa = 10.83SLTVGTTDD1670 pKa = 3.18AAGNAGAPAAPVSLTINTGTGGGGTGGGGTGGGGTGGGSTGGGGTGGDD1718 pKa = 3.8GGTGGSGGSGGSGGTAPVLVGDD1740 pKa = 4.64LNQTLAHH1747 pKa = 6.46APAVSGNLLANDD1759 pKa = 3.74TGAALKK1765 pKa = 8.96VTAVQFSGGLPVAVPTEE1782 pKa = 4.13GTVKK1786 pKa = 10.27VVSDD1790 pKa = 4.02HH1791 pKa = 5.83GTLTVSADD1799 pKa = 3.06GSYY1802 pKa = 10.53SYY1804 pKa = 10.89QAIGANNLDD1813 pKa = 3.67DD1814 pKa = 3.9TVRR1817 pKa = 11.84TAEE1820 pKa = 4.09HH1821 pKa = 5.4FTYY1824 pKa = 10.23TVTDD1828 pKa = 3.28AAGAVGQSSLDD1839 pKa = 3.3ISLGGQAPQASASFGFAFTEE1859 pKa = 4.23ARR1861 pKa = 11.84VTLAGEE1867 pKa = 4.14ALVLVGPDD1875 pKa = 3.04GVTRR1879 pKa = 11.84DD1880 pKa = 3.08ISGIDD1885 pKa = 3.24TLRR1888 pKa = 11.84FTDD1891 pKa = 5.98GEE1893 pKa = 4.1IQNNDD1898 pKa = 2.76GHH1900 pKa = 7.09HH1901 pKa = 5.99VVDD1904 pKa = 5.03DD1905 pKa = 3.66VFYY1908 pKa = 10.62YY1909 pKa = 11.16ANNLDD1914 pKa = 3.19VWRR1917 pKa = 11.84AHH1919 pKa = 6.07VDD1921 pKa = 2.97ADD1923 pKa = 3.56THH1925 pKa = 6.61YY1926 pKa = 9.77EE1927 pKa = 3.98TFGWKK1932 pKa = 9.75EE1933 pKa = 3.67GRR1935 pKa = 11.84DD1936 pKa = 3.63PNAYY1940 pKa = 9.38FHH1942 pKa = 6.25TRR1944 pKa = 11.84AYY1946 pKa = 10.45LAEE1949 pKa = 4.43NPDD1952 pKa = 3.21VAAANINPLEE1962 pKa = 4.59HH1963 pKa = 6.65YY1964 pKa = 8.81LTFGEE1969 pKa = 4.35RR1970 pKa = 11.84EE1971 pKa = 4.18GRR1973 pKa = 11.84SPSPGFSAEE1982 pKa = 4.42AYY1984 pKa = 9.85LHH1986 pKa = 6.6ANPDD1990 pKa = 3.16VAAAGINALAHH2001 pKa = 6.02YY2002 pKa = 7.44LTNGQDD2008 pKa = 3.03EE2009 pKa = 4.65GRR2011 pKa = 11.84TVFAGEE2017 pKa = 4.12SEE2019 pKa = 4.53GPGRR2023 pKa = 11.84VIGDD2027 pKa = 3.85FDD2029 pKa = 5.77ADD2031 pKa = 4.43FYY2033 pKa = 11.05LAQYY2037 pKa = 10.57ADD2039 pKa = 3.38VAAAVPAGRR2048 pKa = 11.84EE2049 pKa = 3.8AASYY2053 pKa = 10.92ALEE2056 pKa = 4.66HH2057 pKa = 6.48YY2058 pKa = 8.73LTFGARR2064 pKa = 11.84EE2065 pKa = 3.95LRR2067 pKa = 11.84NPNAAFDD2074 pKa = 3.65TAYY2077 pKa = 10.97YY2078 pKa = 9.65LQQNPEE2084 pKa = 4.01VAASGANPLLHH2095 pKa = 5.49YY2096 pKa = 10.76QEE2098 pKa = 5.03VGWHH2102 pKa = 5.97EE2103 pKa = 4.01GRR2105 pKa = 11.84NPSASFNTNAYY2116 pKa = 8.95LASHH2120 pKa = 7.15PEE2122 pKa = 3.86VAQAGIDD2129 pKa = 3.77PLEE2132 pKa = 5.33HH2133 pKa = 6.1YY2134 pKa = 10.11LQTGGHH2140 pKa = 6.66ALAA2143 pKa = 5.38
Molecular weight: 205.28 kDa
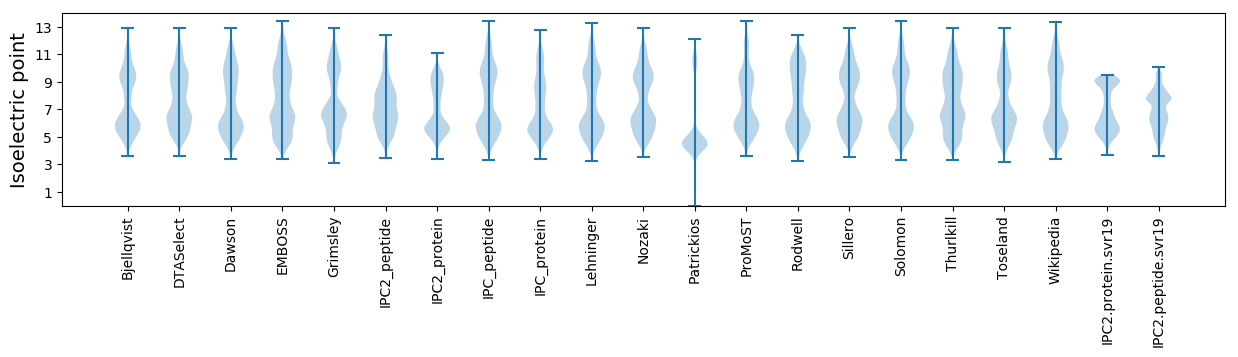
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6MSD0|A0A6N6MSD0_9RHIZ Transglutaminase-like cysteine peptidase OS=Methylobacterium sp. YIM 132548 OX=2615211 GN=F6X51_07385 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84LPLAGLGLFLPLAAWIAAAALVGGSAIATAPGASAQRR39 pKa = 11.84TVTPPAGQPRR49 pKa = 11.84VTAPGGRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84AAHH62 pKa = 5.75HH63 pKa = 6.21HH64 pKa = 6.26RR65 pKa = 11.84PAHH68 pKa = 5.06RR69 pKa = 11.84HH70 pKa = 3.94RR71 pKa = 5.6
MM1 pKa = 7.92RR2 pKa = 11.84LPLAGLGLFLPLAAWIAAAALVGGSAIATAPGASAQRR39 pKa = 11.84TVTPPAGQPRR49 pKa = 11.84VTAPGGRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84AAHH62 pKa = 5.75HH63 pKa = 6.21HH64 pKa = 6.26RR65 pKa = 11.84PAHH68 pKa = 5.06RR69 pKa = 11.84HH70 pKa = 3.94RR71 pKa = 5.6
Molecular weight: 7.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1615088 |
19 |
2478 |
304.5 |
32.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.355 ± 0.056 | 0.821 ± 0.01 |
5.438 ± 0.026 | 5.458 ± 0.031 |
3.369 ± 0.021 | 9.23 ± 0.033 |
1.948 ± 0.015 | 4.491 ± 0.023 |
2.385 ± 0.033 | 10.478 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.008 ± 0.016 | 2.116 ± 0.018 |
5.934 ± 0.034 | 2.769 ± 0.019 |
8.294 ± 0.037 | 4.949 ± 0.027 |
5.299 ± 0.033 | 7.427 ± 0.025 |
1.222 ± 0.014 | 2.009 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
