
Halalkalibacillus sediminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Halalkalibacillus
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
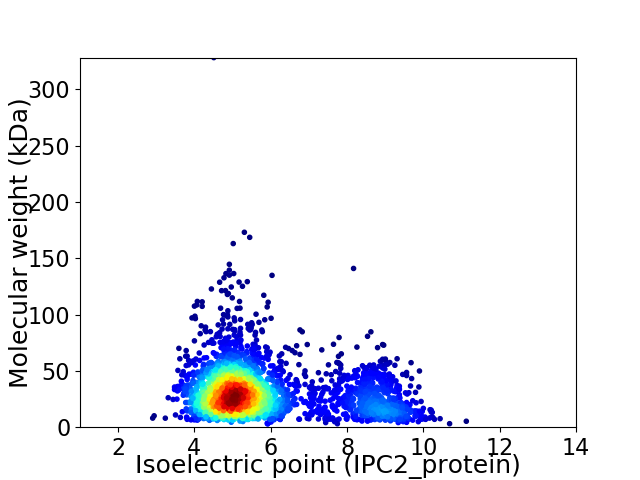
Virtual 2D-PAGE plot for 2899 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
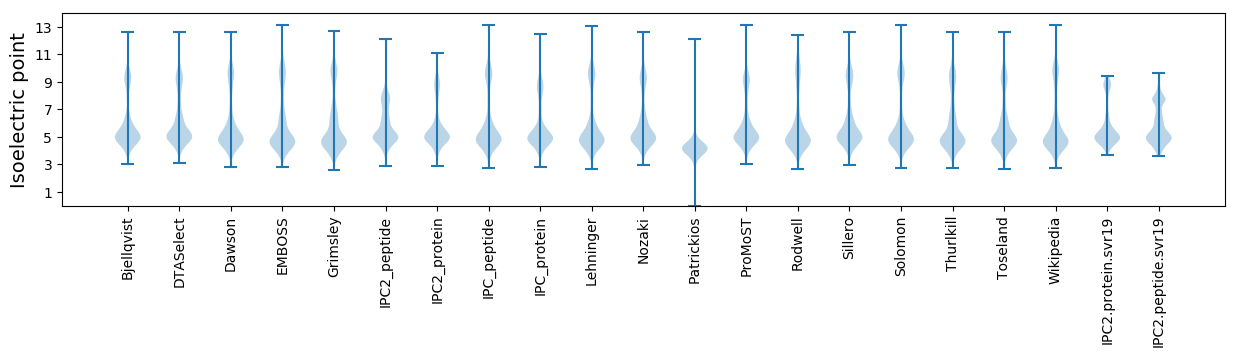
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I0QSI5|A0A2I0QSI5_9BACI Transcriptional regulator OS=Halalkalibacillus sediminis OX=2018042 GN=CEY16_11145 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 10.39NIAVLFTAILLIGLLSACGVLGGGSDD29 pKa = 3.84SDD31 pKa = 4.25SEE33 pKa = 4.65SGGTVTIGGKK43 pKa = 8.94PWTEE47 pKa = 3.93QYY49 pKa = 10.58ILPYY53 pKa = 10.06IIGIHH58 pKa = 5.91LEE60 pKa = 4.14EE61 pKa = 4.37NTDD64 pKa = 3.84FDD66 pKa = 3.81VQYY69 pKa = 11.07EE70 pKa = 4.18EE71 pKa = 4.91GLGEE75 pKa = 4.27VAILTPAIDD84 pKa = 4.93DD85 pKa = 3.88GDD87 pKa = 3.5IDD89 pKa = 4.57VYY91 pKa = 11.33VEE93 pKa = 3.93YY94 pKa = 10.32TGTGLEE100 pKa = 4.12AVLNEE105 pKa = 3.92QAEE108 pKa = 4.55EE109 pKa = 4.55GEE111 pKa = 4.74SADD114 pKa = 3.95SVYY117 pKa = 11.35DD118 pKa = 3.7RR119 pKa = 11.84VKK121 pKa = 10.59EE122 pKa = 4.31GYY124 pKa = 9.65EE125 pKa = 3.78EE126 pKa = 4.18EE127 pKa = 5.59FNVTWLEE134 pKa = 3.74PLGFEE139 pKa = 4.11NTYY142 pKa = 9.21TLAYY146 pKa = 9.79HH147 pKa = 7.15PDD149 pKa = 3.33SGYY152 pKa = 11.13DD153 pKa = 3.22ADD155 pKa = 4.26TYY157 pKa = 11.57SDD159 pKa = 3.69LVEE162 pKa = 4.07TSKK165 pKa = 11.43NEE167 pKa = 3.77DD168 pKa = 3.6MIFGAPHH175 pKa = 6.78AFYY178 pKa = 10.59EE179 pKa = 4.54RR180 pKa = 11.84EE181 pKa = 3.88GDD183 pKa = 3.81GYY185 pKa = 11.29DD186 pKa = 4.18AMLEE190 pKa = 4.12VYY192 pKa = 9.81PFEE195 pKa = 4.16FTEE198 pKa = 4.52TEE200 pKa = 4.13SLDD203 pKa = 3.66ANVMYY208 pKa = 9.9EE209 pKa = 3.83ALEE212 pKa = 4.13SQEE215 pKa = 4.01VDD217 pKa = 4.95LIPAFTTDD225 pKa = 3.92GRR227 pKa = 11.84IEE229 pKa = 4.27RR230 pKa = 11.84YY231 pKa = 9.61NLEE234 pKa = 4.18TTEE237 pKa = 4.6DD238 pKa = 3.66DD239 pKa = 3.33QGFFPKK245 pKa = 10.27YY246 pKa = 9.91DD247 pKa = 3.39AVPLVRR253 pKa = 11.84QEE255 pKa = 4.9ALDD258 pKa = 3.86EE259 pKa = 4.35NPGLKK264 pKa = 9.86EE265 pKa = 4.94AINEE269 pKa = 4.0LAGQISEE276 pKa = 5.02EE277 pKa = 4.19EE278 pKa = 4.07MQEE281 pKa = 3.49MNARR285 pKa = 11.84VDD287 pKa = 3.12IDD289 pKa = 3.81GDD291 pKa = 3.59EE292 pKa = 4.48HH293 pKa = 4.91QTVARR298 pKa = 11.84EE299 pKa = 3.77FLKK302 pKa = 10.84EE303 pKa = 3.83KK304 pKa = 10.83GIIEE308 pKa = 4.14
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 10.39NIAVLFTAILLIGLLSACGVLGGGSDD29 pKa = 3.84SDD31 pKa = 4.25SEE33 pKa = 4.65SGGTVTIGGKK43 pKa = 8.94PWTEE47 pKa = 3.93QYY49 pKa = 10.58ILPYY53 pKa = 10.06IIGIHH58 pKa = 5.91LEE60 pKa = 4.14EE61 pKa = 4.37NTDD64 pKa = 3.84FDD66 pKa = 3.81VQYY69 pKa = 11.07EE70 pKa = 4.18EE71 pKa = 4.91GLGEE75 pKa = 4.27VAILTPAIDD84 pKa = 4.93DD85 pKa = 3.88GDD87 pKa = 3.5IDD89 pKa = 4.57VYY91 pKa = 11.33VEE93 pKa = 3.93YY94 pKa = 10.32TGTGLEE100 pKa = 4.12AVLNEE105 pKa = 3.92QAEE108 pKa = 4.55EE109 pKa = 4.55GEE111 pKa = 4.74SADD114 pKa = 3.95SVYY117 pKa = 11.35DD118 pKa = 3.7RR119 pKa = 11.84VKK121 pKa = 10.59EE122 pKa = 4.31GYY124 pKa = 9.65EE125 pKa = 3.78EE126 pKa = 4.18EE127 pKa = 5.59FNVTWLEE134 pKa = 3.74PLGFEE139 pKa = 4.11NTYY142 pKa = 9.21TLAYY146 pKa = 9.79HH147 pKa = 7.15PDD149 pKa = 3.33SGYY152 pKa = 11.13DD153 pKa = 3.22ADD155 pKa = 4.26TYY157 pKa = 11.57SDD159 pKa = 3.69LVEE162 pKa = 4.07TSKK165 pKa = 11.43NEE167 pKa = 3.77DD168 pKa = 3.6MIFGAPHH175 pKa = 6.78AFYY178 pKa = 10.59EE179 pKa = 4.54RR180 pKa = 11.84EE181 pKa = 3.88GDD183 pKa = 3.81GYY185 pKa = 11.29DD186 pKa = 4.18AMLEE190 pKa = 4.12VYY192 pKa = 9.81PFEE195 pKa = 4.16FTEE198 pKa = 4.52TEE200 pKa = 4.13SLDD203 pKa = 3.66ANVMYY208 pKa = 9.9EE209 pKa = 3.83ALEE212 pKa = 4.13SQEE215 pKa = 4.01VDD217 pKa = 4.95LIPAFTTDD225 pKa = 3.92GRR227 pKa = 11.84IEE229 pKa = 4.27RR230 pKa = 11.84YY231 pKa = 9.61NLEE234 pKa = 4.18TTEE237 pKa = 4.6DD238 pKa = 3.66DD239 pKa = 3.33QGFFPKK245 pKa = 10.27YY246 pKa = 9.91DD247 pKa = 3.39AVPLVRR253 pKa = 11.84QEE255 pKa = 4.9ALDD258 pKa = 3.86EE259 pKa = 4.35NPGLKK264 pKa = 9.86EE265 pKa = 4.94AINEE269 pKa = 4.0LAGQISEE276 pKa = 5.02EE277 pKa = 4.19EE278 pKa = 4.07MQEE281 pKa = 3.49MNARR285 pKa = 11.84VDD287 pKa = 3.12IDD289 pKa = 3.81GDD291 pKa = 3.59EE292 pKa = 4.48HH293 pKa = 4.91QTVARR298 pKa = 11.84EE299 pKa = 3.77FLKK302 pKa = 10.84EE303 pKa = 3.83KK304 pKa = 10.83GIIEE308 pKa = 4.14
Molecular weight: 34.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I0QT15|A0A2I0QT15_9BACI DNA replication initiation control protein YabA OS=Halalkalibacillus sediminis OX=2018042 GN=CEY16_12215 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.28RR12 pKa = 11.84SKK14 pKa = 9.57VHH16 pKa = 5.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.37GRR39 pKa = 11.84KK40 pKa = 6.43TLSAA44 pKa = 4.15
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.28RR12 pKa = 11.84SKK14 pKa = 9.57VHH16 pKa = 5.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.37GRR39 pKa = 11.84KK40 pKa = 6.43TLSAA44 pKa = 4.15
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
832733 |
26 |
2872 |
287.2 |
32.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.318 ± 0.045 | 0.587 ± 0.013 |
5.817 ± 0.042 | 8.419 ± 0.062 |
4.578 ± 0.043 | 6.754 ± 0.044 |
2.131 ± 0.026 | 7.7 ± 0.053 |
6.425 ± 0.047 | 9.402 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.87 ± 0.023 | 4.512 ± 0.035 |
3.478 ± 0.027 | 3.946 ± 0.033 |
4.031 ± 0.035 | 6.226 ± 0.033 |
5.184 ± 0.037 | 7.086 ± 0.033 |
1.003 ± 0.019 | 3.535 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
