
Synechococcus phage S-ShM2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Ahtivirus; Synechococcus virus SShM2
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
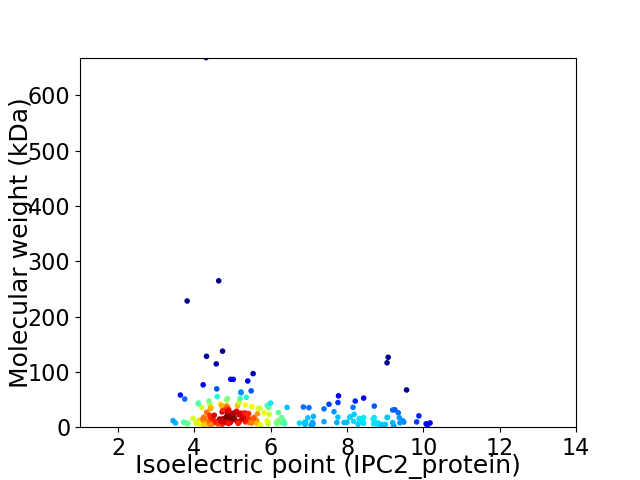
Virtual 2D-PAGE plot for 230 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3SJT4|E3SJT4_9CAUD Uncharacterized protein OS=Synechococcus phage S-ShM2 OX=445683 GN=SShM2_033 PE=4 SV=1
MM1 pKa = 7.28ATQTGKK7 pKa = 10.35LASAKK12 pKa = 9.73PAATTWTALYY22 pKa = 8.84RR23 pKa = 11.84APIDD27 pKa = 3.72SSASGVLNMVSDD39 pKa = 4.32GTAANVRR46 pKa = 11.84VGVKK50 pKa = 10.06KK51 pKa = 10.81YY52 pKa = 10.91DD53 pKa = 3.17IAATLDD59 pKa = 3.14AATYY63 pKa = 9.62LLHH66 pKa = 6.93PGDD69 pKa = 4.27VITNKK74 pKa = 9.28TLTFDD79 pKa = 3.32TSIPIITDD87 pKa = 3.23QQDD90 pKa = 3.19TFTPGQLITTNDD102 pKa = 3.4GEE104 pKa = 4.86TTFKK108 pKa = 10.34WEE110 pKa = 4.1SYY112 pKa = 9.23YY113 pKa = 10.96VPPSTDD119 pKa = 3.27FYY121 pKa = 9.41VQKK124 pKa = 10.57IGILSYY130 pKa = 10.75SIEE133 pKa = 4.11NQTGSFTVGEE143 pKa = 4.51TVTGGTGSGTAVIYY157 pKa = 10.26DD158 pKa = 4.18VIDD161 pKa = 4.14GSLGGSTLYY170 pKa = 10.65LGPVTGTTFVEE181 pKa = 5.09GEE183 pKa = 4.35TLTGGTSSATGDD195 pKa = 3.09ISAGGVGVSRR205 pKa = 11.84DD206 pKa = 3.44EE207 pKa = 5.8LVFSDD212 pKa = 3.63NGSGGTYY219 pKa = 8.96SLRR222 pKa = 11.84RR223 pKa = 11.84STTLTLFLDD232 pKa = 3.69RR233 pKa = 11.84TYY235 pKa = 11.54KK236 pKa = 10.63FFVEE240 pKa = 4.54DD241 pKa = 3.33SSMSGVGFGLSTTINGTFGIDD262 pKa = 3.33QTAGTSDD269 pKa = 4.32DD270 pKa = 3.48GTEE273 pKa = 4.02YY274 pKa = 8.27TTGKK278 pKa = 8.52TSSGTAGSSGAYY290 pKa = 8.54VQYY293 pKa = 11.45DD294 pKa = 3.29MSANGGGDD302 pKa = 2.92ATYY305 pKa = 10.87YY306 pKa = 11.12YY307 pKa = 10.88FDD309 pKa = 4.45TLDD312 pKa = 3.65GTLGGGDD319 pKa = 3.33QSLQLSVDD327 pKa = 3.37YY328 pKa = 10.88SYY330 pKa = 10.68DD331 pKa = 3.24TIYY334 pKa = 10.3IYY336 pKa = 10.72DD337 pKa = 3.96LQGTLTNASDD347 pKa = 5.4AITLGQTTFTLTSNAGSKK365 pKa = 8.42WGYY368 pKa = 7.37VQEE371 pKa = 4.29IDD373 pKa = 3.59GTSVNIVTGLGSADD387 pKa = 3.35FAGADD392 pKa = 3.44TFYY395 pKa = 10.92DD396 pKa = 3.75VPKK399 pKa = 10.21IGTASRR405 pKa = 11.84SLATISSIVTAANAIDD421 pKa = 4.01SVDD424 pKa = 3.25TIMFDD429 pKa = 3.45NSVDD433 pKa = 3.25AAEE436 pKa = 4.64RR437 pKa = 11.84LTSLVIGPGQVLMVYY452 pKa = 10.24AATQNISFDD461 pKa = 3.46YY462 pKa = 11.21SGFQDD467 pKa = 3.21SSSDD471 pKa = 3.41FTVRR475 pKa = 11.84SFDD478 pKa = 3.66VNASIAADD486 pKa = 3.87GASGGGG492 pKa = 3.32
MM1 pKa = 7.28ATQTGKK7 pKa = 10.35LASAKK12 pKa = 9.73PAATTWTALYY22 pKa = 8.84RR23 pKa = 11.84APIDD27 pKa = 3.72SSASGVLNMVSDD39 pKa = 4.32GTAANVRR46 pKa = 11.84VGVKK50 pKa = 10.06KK51 pKa = 10.81YY52 pKa = 10.91DD53 pKa = 3.17IAATLDD59 pKa = 3.14AATYY63 pKa = 9.62LLHH66 pKa = 6.93PGDD69 pKa = 4.27VITNKK74 pKa = 9.28TLTFDD79 pKa = 3.32TSIPIITDD87 pKa = 3.23QQDD90 pKa = 3.19TFTPGQLITTNDD102 pKa = 3.4GEE104 pKa = 4.86TTFKK108 pKa = 10.34WEE110 pKa = 4.1SYY112 pKa = 9.23YY113 pKa = 10.96VPPSTDD119 pKa = 3.27FYY121 pKa = 9.41VQKK124 pKa = 10.57IGILSYY130 pKa = 10.75SIEE133 pKa = 4.11NQTGSFTVGEE143 pKa = 4.51TVTGGTGSGTAVIYY157 pKa = 10.26DD158 pKa = 4.18VIDD161 pKa = 4.14GSLGGSTLYY170 pKa = 10.65LGPVTGTTFVEE181 pKa = 5.09GEE183 pKa = 4.35TLTGGTSSATGDD195 pKa = 3.09ISAGGVGVSRR205 pKa = 11.84DD206 pKa = 3.44EE207 pKa = 5.8LVFSDD212 pKa = 3.63NGSGGTYY219 pKa = 8.96SLRR222 pKa = 11.84RR223 pKa = 11.84STTLTLFLDD232 pKa = 3.69RR233 pKa = 11.84TYY235 pKa = 11.54KK236 pKa = 10.63FFVEE240 pKa = 4.54DD241 pKa = 3.33SSMSGVGFGLSTTINGTFGIDD262 pKa = 3.33QTAGTSDD269 pKa = 4.32DD270 pKa = 3.48GTEE273 pKa = 4.02YY274 pKa = 8.27TTGKK278 pKa = 8.52TSSGTAGSSGAYY290 pKa = 8.54VQYY293 pKa = 11.45DD294 pKa = 3.29MSANGGGDD302 pKa = 2.92ATYY305 pKa = 10.87YY306 pKa = 11.12YY307 pKa = 10.88FDD309 pKa = 4.45TLDD312 pKa = 3.65GTLGGGDD319 pKa = 3.33QSLQLSVDD327 pKa = 3.37YY328 pKa = 10.88SYY330 pKa = 10.68DD331 pKa = 3.24TIYY334 pKa = 10.3IYY336 pKa = 10.72DD337 pKa = 3.96LQGTLTNASDD347 pKa = 5.4AITLGQTTFTLTSNAGSKK365 pKa = 8.42WGYY368 pKa = 7.37VQEE371 pKa = 4.29IDD373 pKa = 3.59GTSVNIVTGLGSADD387 pKa = 3.35FAGADD392 pKa = 3.44TFYY395 pKa = 10.92DD396 pKa = 3.75VPKK399 pKa = 10.21IGTASRR405 pKa = 11.84SLATISSIVTAANAIDD421 pKa = 4.01SVDD424 pKa = 3.25TIMFDD429 pKa = 3.45NSVDD433 pKa = 3.25AAEE436 pKa = 4.64RR437 pKa = 11.84LTSLVIGPGQVLMVYY452 pKa = 10.24AATQNISFDD461 pKa = 3.46YY462 pKa = 11.21SGFQDD467 pKa = 3.21SSSDD471 pKa = 3.41FTVRR475 pKa = 11.84SFDD478 pKa = 3.66VNASIAADD486 pKa = 3.87GASGGGG492 pKa = 3.32
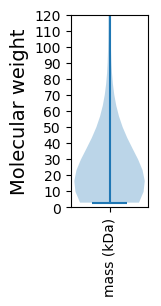
Molecular weight: 50.91 kDa
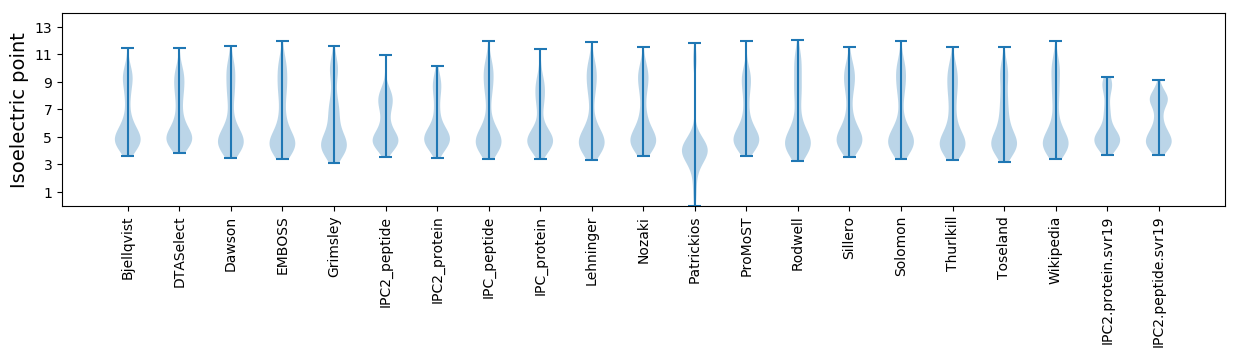
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3SJQ9|E3SJQ9_9CAUD Head completion protein OS=Synechococcus phage S-ShM2 OX=445683 GN=gp4 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.37KK3 pKa = 10.28VLLLIAGAAFFAAPVQAHH21 pKa = 5.31QAHH24 pKa = 6.88KK25 pKa = 10.27HH26 pKa = 5.54RR27 pKa = 11.84NHH29 pKa = 5.97HH30 pKa = 4.92HH31 pKa = 6.77HH32 pKa = 6.33YY33 pKa = 10.72NNVEE37 pKa = 4.07RR38 pKa = 11.84YY39 pKa = 6.93FTCHH43 pKa = 5.63DD44 pKa = 3.44HH45 pKa = 6.24LRR47 pKa = 11.84RR48 pKa = 11.84NIRR51 pKa = 11.84HH52 pKa = 5.71CHH54 pKa = 4.07GHH56 pKa = 5.26TNWGHH61 pKa = 4.9GRR63 pKa = 11.84QLRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84DD69 pKa = 3.47YY70 pKa = 9.66FAPSIIFDD78 pKa = 3.81FF79 pKa = 4.75
MM1 pKa = 7.5KK2 pKa = 10.37KK3 pKa = 10.28VLLLIAGAAFFAAPVQAHH21 pKa = 5.31QAHH24 pKa = 6.88KK25 pKa = 10.27HH26 pKa = 5.54RR27 pKa = 11.84NHH29 pKa = 5.97HH30 pKa = 4.92HH31 pKa = 6.77HH32 pKa = 6.33YY33 pKa = 10.72NNVEE37 pKa = 4.07RR38 pKa = 11.84YY39 pKa = 6.93FTCHH43 pKa = 5.63DD44 pKa = 3.44HH45 pKa = 6.24LRR47 pKa = 11.84RR48 pKa = 11.84NIRR51 pKa = 11.84HH52 pKa = 5.71CHH54 pKa = 4.07GHH56 pKa = 5.26TNWGHH61 pKa = 4.9GRR63 pKa = 11.84QLRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84DD69 pKa = 3.47YY70 pKa = 9.66FAPSIIFDD78 pKa = 3.81FF79 pKa = 4.75
Molecular weight: 9.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
58486 |
24 |
6198 |
254.3 |
28.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.841 ± 0.214 | 0.983 ± 0.089 |
6.737 ± 0.145 | 5.995 ± 0.268 |
4.283 ± 0.091 | 7.904 ± 0.346 |
1.477 ± 0.105 | 6.319 ± 0.202 |
5.658 ± 0.406 | 7.099 ± 0.148 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.193 | 5.701 ± 0.175 |
4.045 ± 0.149 | 3.85 ± 0.096 |
4.275 ± 0.169 | 7.185 ± 0.165 |
7.34 ± 0.397 | 6.615 ± 0.176 |
1.185 ± 0.09 | 4.259 ± 0.139 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
