
Hungateiclostridiaceae bacterium KB18
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; unclassified Oscillospiraceae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
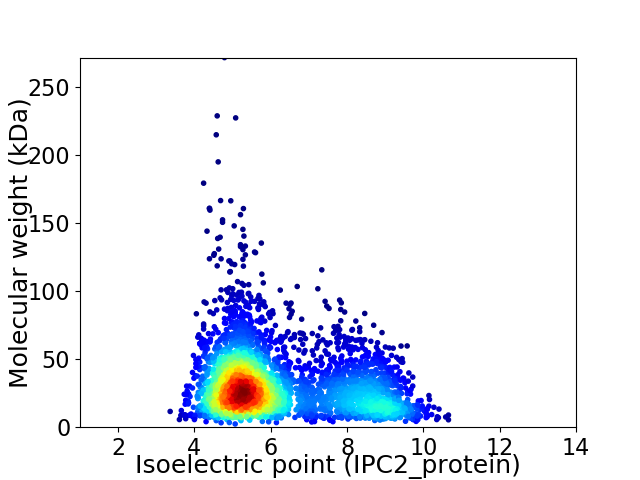
Virtual 2D-PAGE plot for 3593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C7GNG5|A0A1C7GNG5_9FIRM ABC transporter permease OS=Hungateiclostridiaceae bacterium KB18 OX=1834198 GN=A4V00_03700 PE=4 SV=1
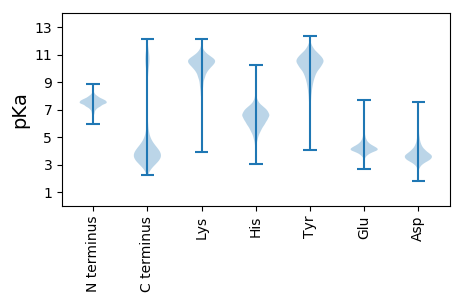
MM1 pKa = 7.6KK2 pKa = 10.47KK3 pKa = 10.39LLALVLALCLVLGLGGCRR21 pKa = 11.84FINKK25 pKa = 6.24TTAPVEE31 pKa = 4.23FTSADD36 pKa = 3.29SGTLYY41 pKa = 8.68WHH43 pKa = 6.22SMEE46 pKa = 4.74LAGLIDD52 pKa = 5.69DD53 pKa = 4.07MLKK56 pKa = 8.67TPEE59 pKa = 4.13YY60 pKa = 9.52FEE62 pKa = 5.74LMGGSDD68 pKa = 3.62TVSEE72 pKa = 4.42VIIPLMRR79 pKa = 11.84SDD81 pKa = 3.69LSEE84 pKa = 4.4PEE86 pKa = 3.76SAYY89 pKa = 11.01VVTLPEE95 pKa = 4.64DD96 pKa = 3.82ALPTLMSAAEE106 pKa = 3.89VDD108 pKa = 3.71LDD110 pKa = 3.94GFSDD114 pKa = 4.09GLRR117 pKa = 11.84EE118 pKa = 3.99FVEE121 pKa = 4.15RR122 pKa = 11.84RR123 pKa = 11.84MASSVPTILNSRR135 pKa = 11.84QGAEE139 pKa = 3.89TLAASSILTVSGSWAGEE156 pKa = 3.96VLDD159 pKa = 4.38EE160 pKa = 4.26SCYY163 pKa = 10.49VLLMYY168 pKa = 8.04PYY170 pKa = 9.59CCPVMASFTGSNGVVTGTATMLLGEE195 pKa = 4.46PLEE198 pKa = 4.5EE199 pKa = 5.22DD200 pKa = 3.2SLDD203 pKa = 3.6AVEE206 pKa = 5.18EE207 pKa = 4.12LFGYY211 pKa = 10.9LEE213 pKa = 4.13MEE215 pKa = 4.31GPEE218 pKa = 4.08IEE220 pKa = 5.25EE221 pKa = 3.86IDD223 pKa = 3.46IDD225 pKa = 4.41ALII228 pKa = 4.48
MM1 pKa = 7.6KK2 pKa = 10.47KK3 pKa = 10.39LLALVLALCLVLGLGGCRR21 pKa = 11.84FINKK25 pKa = 6.24TTAPVEE31 pKa = 4.23FTSADD36 pKa = 3.29SGTLYY41 pKa = 8.68WHH43 pKa = 6.22SMEE46 pKa = 4.74LAGLIDD52 pKa = 5.69DD53 pKa = 4.07MLKK56 pKa = 8.67TPEE59 pKa = 4.13YY60 pKa = 9.52FEE62 pKa = 5.74LMGGSDD68 pKa = 3.62TVSEE72 pKa = 4.42VIIPLMRR79 pKa = 11.84SDD81 pKa = 3.69LSEE84 pKa = 4.4PEE86 pKa = 3.76SAYY89 pKa = 11.01VVTLPEE95 pKa = 4.64DD96 pKa = 3.82ALPTLMSAAEE106 pKa = 3.89VDD108 pKa = 3.71LDD110 pKa = 3.94GFSDD114 pKa = 4.09GLRR117 pKa = 11.84EE118 pKa = 3.99FVEE121 pKa = 4.15RR122 pKa = 11.84RR123 pKa = 11.84MASSVPTILNSRR135 pKa = 11.84QGAEE139 pKa = 3.89TLAASSILTVSGSWAGEE156 pKa = 3.96VLDD159 pKa = 4.38EE160 pKa = 4.26SCYY163 pKa = 10.49VLLMYY168 pKa = 8.04PYY170 pKa = 9.59CCPVMASFTGSNGVVTGTATMLLGEE195 pKa = 4.46PLEE198 pKa = 4.5EE199 pKa = 5.22DD200 pKa = 3.2SLDD203 pKa = 3.6AVEE206 pKa = 5.18EE207 pKa = 4.12LFGYY211 pKa = 10.9LEE213 pKa = 4.13MEE215 pKa = 4.31GPEE218 pKa = 4.08IEE220 pKa = 5.25EE221 pKa = 3.86IDD223 pKa = 3.46IDD225 pKa = 4.41ALII228 pKa = 4.48
Molecular weight: 24.56 kDa
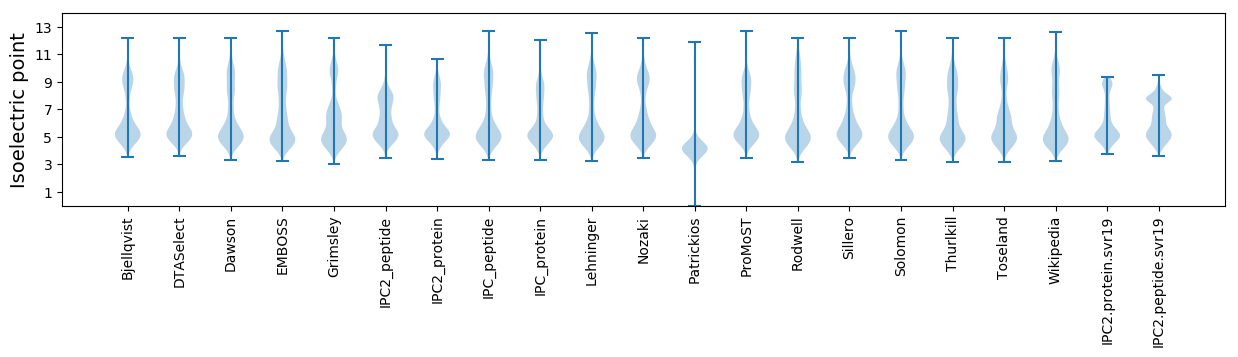
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0QDZ3|A0A1V0QDZ3_9FIRM Uncharacterized protein OS=Hungateiclostridiaceae bacterium KB18 OX=1834198 GN=A4V00_19230 PE=4 SV=1
MM1 pKa = 7.53HH2 pKa = 7.92PLHH5 pKa = 6.58LQLPPLEE12 pKa = 4.47EE13 pKa = 3.99LHH15 pKa = 6.74LLRR18 pKa = 11.84PSGAGVRR25 pKa = 11.84RR26 pKa = 11.84QGDD29 pKa = 3.98LRR31 pKa = 11.84FMLCEE36 pKa = 3.79ASINQQSLTQFMDD49 pKa = 3.29RR50 pKa = 11.84LVRR53 pKa = 11.84TSLRR57 pKa = 11.84KK58 pKa = 9.95VFLILDD64 pKa = 3.83NLKK67 pKa = 10.19GHH69 pKa = 6.77HH70 pKa = 6.26GKK72 pKa = 9.97KK73 pKa = 9.9AAARR77 pKa = 11.84LDD79 pKa = 3.42RR80 pKa = 11.84HH81 pKa = 5.83RR82 pKa = 11.84KK83 pKa = 9.41KK84 pKa = 10.53IEE86 pKa = 3.89LFLLAAYY93 pKa = 8.74LRR95 pKa = 11.84PNVISAIKK103 pKa = 9.33PPPSCEE109 pKa = 4.07PYY111 pKa = 9.89NLLPIRR117 pKa = 11.84SPLSFCMIRR126 pKa = 11.84FPVFLFGGNTPPEE139 pKa = 4.73DD140 pKa = 3.27GFAPQGRR147 pKa = 11.84GCVYY151 pKa = 10.72LSGRR155 pKa = 11.84PGKK158 pKa = 10.58SGGRR162 pKa = 11.84VLTCHH167 pKa = 6.15CQGPLRR173 pKa = 11.84SLGGRR178 pKa = 11.84RR179 pKa = 11.84LLSS182 pKa = 3.49
MM1 pKa = 7.53HH2 pKa = 7.92PLHH5 pKa = 6.58LQLPPLEE12 pKa = 4.47EE13 pKa = 3.99LHH15 pKa = 6.74LLRR18 pKa = 11.84PSGAGVRR25 pKa = 11.84RR26 pKa = 11.84QGDD29 pKa = 3.98LRR31 pKa = 11.84FMLCEE36 pKa = 3.79ASINQQSLTQFMDD49 pKa = 3.29RR50 pKa = 11.84LVRR53 pKa = 11.84TSLRR57 pKa = 11.84KK58 pKa = 9.95VFLILDD64 pKa = 3.83NLKK67 pKa = 10.19GHH69 pKa = 6.77HH70 pKa = 6.26GKK72 pKa = 9.97KK73 pKa = 9.9AAARR77 pKa = 11.84LDD79 pKa = 3.42RR80 pKa = 11.84HH81 pKa = 5.83RR82 pKa = 11.84KK83 pKa = 9.41KK84 pKa = 10.53IEE86 pKa = 3.89LFLLAAYY93 pKa = 8.74LRR95 pKa = 11.84PNVISAIKK103 pKa = 9.33PPPSCEE109 pKa = 4.07PYY111 pKa = 9.89NLLPIRR117 pKa = 11.84SPLSFCMIRR126 pKa = 11.84FPVFLFGGNTPPEE139 pKa = 4.73DD140 pKa = 3.27GFAPQGRR147 pKa = 11.84GCVYY151 pKa = 10.72LSGRR155 pKa = 11.84PGKK158 pKa = 10.58SGGRR162 pKa = 11.84VLTCHH167 pKa = 6.15CQGPLRR173 pKa = 11.84SLGGRR178 pKa = 11.84RR179 pKa = 11.84LLSS182 pKa = 3.49
Molecular weight: 20.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066074 |
22 |
2513 |
296.7 |
33.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.456 ± 0.044 | 1.619 ± 0.018 |
5.491 ± 0.03 | 7.402 ± 0.043 |
4.006 ± 0.025 | 7.862 ± 0.044 |
1.749 ± 0.017 | 5.768 ± 0.033 |
5.4 ± 0.043 | 9.797 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.854 ± 0.02 | 3.595 ± 0.028 |
4.245 ± 0.03 | 3.313 ± 0.025 |
5.699 ± 0.048 | 5.941 ± 0.033 |
5.245 ± 0.042 | 6.747 ± 0.033 |
1.107 ± 0.015 | 3.705 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
