
Lake Sarah-associated circular molecule 3
Taxonomy: Viruses; unclassified viruses
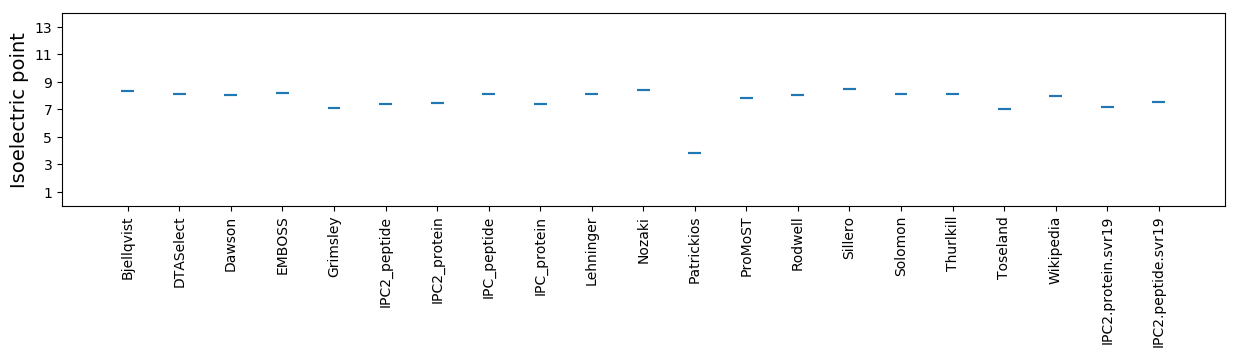
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G8F9|A0A126G8F9_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 3 OX=1685728 PE=3 SV=1
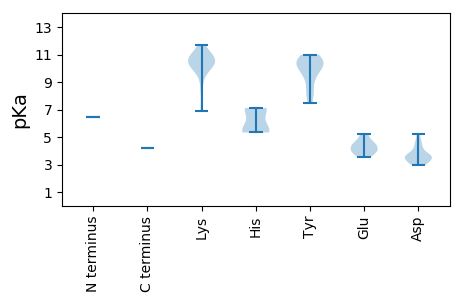
MM1 pKa = 6.48YY2 pKa = 7.91TNPNSYY8 pKa = 10.8FDD10 pKa = 3.42LTLPQRR16 pKa = 11.84RR17 pKa = 11.84FMFTSYY23 pKa = 9.73TLEE26 pKa = 4.56GEE28 pKa = 4.55GFTVPDD34 pKa = 3.05QDD36 pKa = 3.14PRR38 pKa = 11.84FRR40 pKa = 11.84YY41 pKa = 9.24CVYY44 pKa = 10.28QAEE47 pKa = 4.74KK48 pKa = 10.57CPQTGNIHH56 pKa = 5.34LQGYY60 pKa = 9.55IEE62 pKa = 5.08FKK64 pKa = 10.85DD65 pKa = 3.6QIRR68 pKa = 11.84GNTVQKK74 pKa = 10.82LMNVPGLHH82 pKa = 7.05LSRR85 pKa = 11.84VDD87 pKa = 3.63YY88 pKa = 10.49PEE90 pKa = 4.2KK91 pKa = 10.77ARR93 pKa = 11.84AYY95 pKa = 10.46CMKK98 pKa = 10.35LDD100 pKa = 3.52TRR102 pKa = 11.84YY103 pKa = 10.54AEE105 pKa = 3.98PQEE108 pKa = 3.75YY109 pKa = 10.47GKK111 pKa = 10.72YY112 pKa = 9.98KK113 pKa = 10.5KK114 pKa = 10.2EE115 pKa = 3.52QGARR119 pKa = 11.84NDD121 pKa = 3.77LKK123 pKa = 11.69AMGDD127 pKa = 3.59MVLAGATHH135 pKa = 5.77IQIYY139 pKa = 9.81DD140 pKa = 4.18AYY142 pKa = 8.14PGQYY146 pKa = 8.81IRR148 pKa = 11.84YY149 pKa = 8.44YY150 pKa = 10.51KK151 pKa = 10.71AIDD154 pKa = 3.64RR155 pKa = 11.84LRR157 pKa = 11.84QIYY160 pKa = 10.61ALDD163 pKa = 3.47KK164 pKa = 10.68PNRR167 pKa = 11.84LKK169 pKa = 10.41IGRR172 pKa = 11.84MEE174 pKa = 5.23CILFYY179 pKa = 10.97GRR181 pKa = 11.84PGTGKK186 pKa = 6.92TTQAKK191 pKa = 9.66EE192 pKa = 3.72YY193 pKa = 10.07AAHH196 pKa = 7.11IKK198 pKa = 8.5QAYY201 pKa = 7.45WEE203 pKa = 4.18IPIANSGTLWFDD215 pKa = 3.78GYY217 pKa = 10.98NAEE220 pKa = 4.25PCVIIDD226 pKa = 4.52DD227 pKa = 4.36FSGKK231 pKa = 10.02INLDD235 pKa = 2.99AALRR239 pKa = 11.84IFDD242 pKa = 3.59QGEE245 pKa = 3.94VQVPVKK251 pKa = 10.43GGHH254 pKa = 5.41VWWCPKK260 pKa = 10.03TIIITSNVHH269 pKa = 5.33PSNWYY274 pKa = 9.93DD275 pKa = 3.42YY276 pKa = 10.68QNRR279 pKa = 11.84QDD281 pKa = 4.75SMIAMYY287 pKa = 10.29RR288 pKa = 11.84RR289 pKa = 11.84LHH291 pKa = 7.05RR292 pKa = 11.84IVCFNRR298 pKa = 11.84ATPAEE303 pKa = 3.91VDD305 pKa = 3.14RR306 pKa = 11.84FKK308 pKa = 11.3FLDD311 pKa = 3.46IPEE314 pKa = 4.33NAFDD318 pKa = 5.2VIMEE322 pKa = 4.51TDD324 pKa = 2.97QQ325 pKa = 4.18
MM1 pKa = 6.48YY2 pKa = 7.91TNPNSYY8 pKa = 10.8FDD10 pKa = 3.42LTLPQRR16 pKa = 11.84RR17 pKa = 11.84FMFTSYY23 pKa = 9.73TLEE26 pKa = 4.56GEE28 pKa = 4.55GFTVPDD34 pKa = 3.05QDD36 pKa = 3.14PRR38 pKa = 11.84FRR40 pKa = 11.84YY41 pKa = 9.24CVYY44 pKa = 10.28QAEE47 pKa = 4.74KK48 pKa = 10.57CPQTGNIHH56 pKa = 5.34LQGYY60 pKa = 9.55IEE62 pKa = 5.08FKK64 pKa = 10.85DD65 pKa = 3.6QIRR68 pKa = 11.84GNTVQKK74 pKa = 10.82LMNVPGLHH82 pKa = 7.05LSRR85 pKa = 11.84VDD87 pKa = 3.63YY88 pKa = 10.49PEE90 pKa = 4.2KK91 pKa = 10.77ARR93 pKa = 11.84AYY95 pKa = 10.46CMKK98 pKa = 10.35LDD100 pKa = 3.52TRR102 pKa = 11.84YY103 pKa = 10.54AEE105 pKa = 3.98PQEE108 pKa = 3.75YY109 pKa = 10.47GKK111 pKa = 10.72YY112 pKa = 9.98KK113 pKa = 10.5KK114 pKa = 10.2EE115 pKa = 3.52QGARR119 pKa = 11.84NDD121 pKa = 3.77LKK123 pKa = 11.69AMGDD127 pKa = 3.59MVLAGATHH135 pKa = 5.77IQIYY139 pKa = 9.81DD140 pKa = 4.18AYY142 pKa = 8.14PGQYY146 pKa = 8.81IRR148 pKa = 11.84YY149 pKa = 8.44YY150 pKa = 10.51KK151 pKa = 10.71AIDD154 pKa = 3.64RR155 pKa = 11.84LRR157 pKa = 11.84QIYY160 pKa = 10.61ALDD163 pKa = 3.47KK164 pKa = 10.68PNRR167 pKa = 11.84LKK169 pKa = 10.41IGRR172 pKa = 11.84MEE174 pKa = 5.23CILFYY179 pKa = 10.97GRR181 pKa = 11.84PGTGKK186 pKa = 6.92TTQAKK191 pKa = 9.66EE192 pKa = 3.72YY193 pKa = 10.07AAHH196 pKa = 7.11IKK198 pKa = 8.5QAYY201 pKa = 7.45WEE203 pKa = 4.18IPIANSGTLWFDD215 pKa = 3.78GYY217 pKa = 10.98NAEE220 pKa = 4.25PCVIIDD226 pKa = 4.52DD227 pKa = 4.36FSGKK231 pKa = 10.02INLDD235 pKa = 2.99AALRR239 pKa = 11.84IFDD242 pKa = 3.59QGEE245 pKa = 3.94VQVPVKK251 pKa = 10.43GGHH254 pKa = 5.41VWWCPKK260 pKa = 10.03TIIITSNVHH269 pKa = 5.33PSNWYY274 pKa = 9.93DD275 pKa = 3.42YY276 pKa = 10.68QNRR279 pKa = 11.84QDD281 pKa = 4.75SMIAMYY287 pKa = 10.29RR288 pKa = 11.84RR289 pKa = 11.84LHH291 pKa = 7.05RR292 pKa = 11.84IVCFNRR298 pKa = 11.84ATPAEE303 pKa = 3.91VDD305 pKa = 3.14RR306 pKa = 11.84FKK308 pKa = 11.3FLDD311 pKa = 3.46IPEE314 pKa = 4.33NAFDD318 pKa = 5.2VIMEE322 pKa = 4.51TDD324 pKa = 2.97QQ325 pKa = 4.18
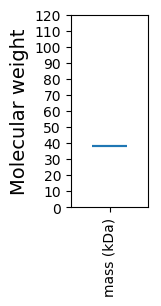
Molecular weight: 37.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G8F9|A0A126G8F9_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 3 OX=1685728 PE=3 SV=1
MM1 pKa = 6.48YY2 pKa = 7.91TNPNSYY8 pKa = 10.8FDD10 pKa = 3.42LTLPQRR16 pKa = 11.84RR17 pKa = 11.84FMFTSYY23 pKa = 9.73TLEE26 pKa = 4.56GEE28 pKa = 4.55GFTVPDD34 pKa = 3.05QDD36 pKa = 3.14PRR38 pKa = 11.84FRR40 pKa = 11.84YY41 pKa = 9.24CVYY44 pKa = 10.28QAEE47 pKa = 4.74KK48 pKa = 10.57CPQTGNIHH56 pKa = 5.34LQGYY60 pKa = 9.55IEE62 pKa = 5.08FKK64 pKa = 10.85DD65 pKa = 3.6QIRR68 pKa = 11.84GNTVQKK74 pKa = 10.82LMNVPGLHH82 pKa = 7.05LSRR85 pKa = 11.84VDD87 pKa = 3.63YY88 pKa = 10.49PEE90 pKa = 4.2KK91 pKa = 10.77ARR93 pKa = 11.84AYY95 pKa = 10.46CMKK98 pKa = 10.35LDD100 pKa = 3.52TRR102 pKa = 11.84YY103 pKa = 10.54AEE105 pKa = 3.98PQEE108 pKa = 3.75YY109 pKa = 10.47GKK111 pKa = 10.72YY112 pKa = 9.98KK113 pKa = 10.5KK114 pKa = 10.2EE115 pKa = 3.52QGARR119 pKa = 11.84NDD121 pKa = 3.77LKK123 pKa = 11.69AMGDD127 pKa = 3.59MVLAGATHH135 pKa = 5.77IQIYY139 pKa = 9.81DD140 pKa = 4.18AYY142 pKa = 8.14PGQYY146 pKa = 8.81IRR148 pKa = 11.84YY149 pKa = 8.44YY150 pKa = 10.51KK151 pKa = 10.71AIDD154 pKa = 3.64RR155 pKa = 11.84LRR157 pKa = 11.84QIYY160 pKa = 10.61ALDD163 pKa = 3.47KK164 pKa = 10.68PNRR167 pKa = 11.84LKK169 pKa = 10.41IGRR172 pKa = 11.84MEE174 pKa = 5.23CILFYY179 pKa = 10.97GRR181 pKa = 11.84PGTGKK186 pKa = 6.92TTQAKK191 pKa = 9.66EE192 pKa = 3.72YY193 pKa = 10.07AAHH196 pKa = 7.11IKK198 pKa = 8.5QAYY201 pKa = 7.45WEE203 pKa = 4.18IPIANSGTLWFDD215 pKa = 3.78GYY217 pKa = 10.98NAEE220 pKa = 4.25PCVIIDD226 pKa = 4.52DD227 pKa = 4.36FSGKK231 pKa = 10.02INLDD235 pKa = 2.99AALRR239 pKa = 11.84IFDD242 pKa = 3.59QGEE245 pKa = 3.94VQVPVKK251 pKa = 10.43GGHH254 pKa = 5.41VWWCPKK260 pKa = 10.03TIIITSNVHH269 pKa = 5.33PSNWYY274 pKa = 9.93DD275 pKa = 3.42YY276 pKa = 10.68QNRR279 pKa = 11.84QDD281 pKa = 4.75SMIAMYY287 pKa = 10.29RR288 pKa = 11.84RR289 pKa = 11.84LHH291 pKa = 7.05RR292 pKa = 11.84IVCFNRR298 pKa = 11.84ATPAEE303 pKa = 3.91VDD305 pKa = 3.14RR306 pKa = 11.84FKK308 pKa = 11.3FLDD311 pKa = 3.46IPEE314 pKa = 4.33NAFDD318 pKa = 5.2VIMEE322 pKa = 4.51TDD324 pKa = 2.97QQ325 pKa = 4.18
MM1 pKa = 6.48YY2 pKa = 7.91TNPNSYY8 pKa = 10.8FDD10 pKa = 3.42LTLPQRR16 pKa = 11.84RR17 pKa = 11.84FMFTSYY23 pKa = 9.73TLEE26 pKa = 4.56GEE28 pKa = 4.55GFTVPDD34 pKa = 3.05QDD36 pKa = 3.14PRR38 pKa = 11.84FRR40 pKa = 11.84YY41 pKa = 9.24CVYY44 pKa = 10.28QAEE47 pKa = 4.74KK48 pKa = 10.57CPQTGNIHH56 pKa = 5.34LQGYY60 pKa = 9.55IEE62 pKa = 5.08FKK64 pKa = 10.85DD65 pKa = 3.6QIRR68 pKa = 11.84GNTVQKK74 pKa = 10.82LMNVPGLHH82 pKa = 7.05LSRR85 pKa = 11.84VDD87 pKa = 3.63YY88 pKa = 10.49PEE90 pKa = 4.2KK91 pKa = 10.77ARR93 pKa = 11.84AYY95 pKa = 10.46CMKK98 pKa = 10.35LDD100 pKa = 3.52TRR102 pKa = 11.84YY103 pKa = 10.54AEE105 pKa = 3.98PQEE108 pKa = 3.75YY109 pKa = 10.47GKK111 pKa = 10.72YY112 pKa = 9.98KK113 pKa = 10.5KK114 pKa = 10.2EE115 pKa = 3.52QGARR119 pKa = 11.84NDD121 pKa = 3.77LKK123 pKa = 11.69AMGDD127 pKa = 3.59MVLAGATHH135 pKa = 5.77IQIYY139 pKa = 9.81DD140 pKa = 4.18AYY142 pKa = 8.14PGQYY146 pKa = 8.81IRR148 pKa = 11.84YY149 pKa = 8.44YY150 pKa = 10.51KK151 pKa = 10.71AIDD154 pKa = 3.64RR155 pKa = 11.84LRR157 pKa = 11.84QIYY160 pKa = 10.61ALDD163 pKa = 3.47KK164 pKa = 10.68PNRR167 pKa = 11.84LKK169 pKa = 10.41IGRR172 pKa = 11.84MEE174 pKa = 5.23CILFYY179 pKa = 10.97GRR181 pKa = 11.84PGTGKK186 pKa = 6.92TTQAKK191 pKa = 9.66EE192 pKa = 3.72YY193 pKa = 10.07AAHH196 pKa = 7.11IKK198 pKa = 8.5QAYY201 pKa = 7.45WEE203 pKa = 4.18IPIANSGTLWFDD215 pKa = 3.78GYY217 pKa = 10.98NAEE220 pKa = 4.25PCVIIDD226 pKa = 4.52DD227 pKa = 4.36FSGKK231 pKa = 10.02INLDD235 pKa = 2.99AALRR239 pKa = 11.84IFDD242 pKa = 3.59QGEE245 pKa = 3.94VQVPVKK251 pKa = 10.43GGHH254 pKa = 5.41VWWCPKK260 pKa = 10.03TIIITSNVHH269 pKa = 5.33PSNWYY274 pKa = 9.93DD275 pKa = 3.42YY276 pKa = 10.68QNRR279 pKa = 11.84QDD281 pKa = 4.75SMIAMYY287 pKa = 10.29RR288 pKa = 11.84RR289 pKa = 11.84LHH291 pKa = 7.05RR292 pKa = 11.84IVCFNRR298 pKa = 11.84ATPAEE303 pKa = 3.91VDD305 pKa = 3.14RR306 pKa = 11.84FKK308 pKa = 11.3FLDD311 pKa = 3.46IPEE314 pKa = 4.33NAFDD318 pKa = 5.2VIMEE322 pKa = 4.51TDD324 pKa = 2.97QQ325 pKa = 4.18
Molecular weight: 37.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
325 |
325 |
325 |
325.0 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.077 ± 0.0 | 2.154 ± 0.0 |
6.769 ± 0.0 | 4.923 ± 0.0 |
4.308 ± 0.0 | 6.462 ± 0.0 |
2.154 ± 0.0 | 7.385 ± 0.0 |
5.846 ± 0.0 | 5.846 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.0 | 4.615 ± 0.0 |
5.538 ± 0.0 | 5.846 ± 0.0 |
6.769 ± 0.0 | 2.462 ± 0.0 |
5.231 ± 0.0 | 4.615 ± 0.0 |
1.538 ± 0.0 | 7.385 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
