
Wuhan cricket virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
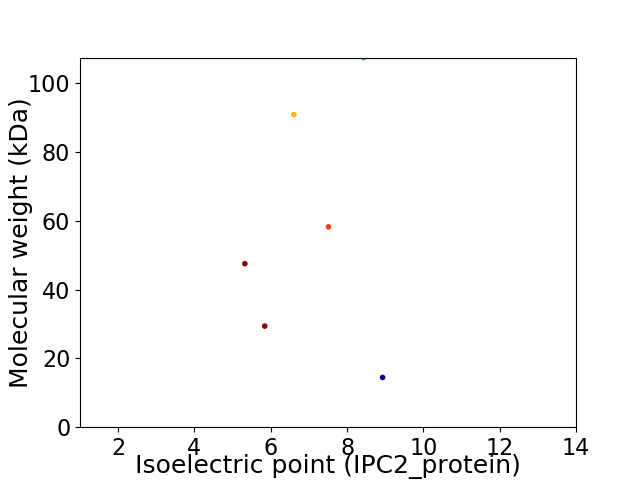
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0QKT8|A0A0P0QKT8_9VIRU Putative glycoprotein OS=Wuhan cricket virus OX=1746070 GN=VP1 PE=4 SV=1
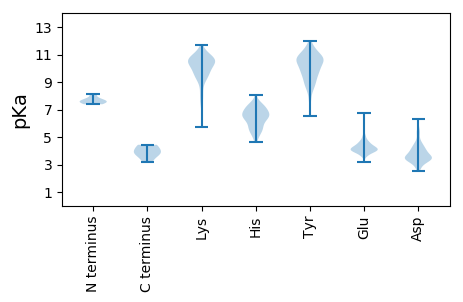
MM1 pKa = 7.58GSYY4 pKa = 8.83WCSNFALLWVQKK16 pKa = 9.93RR17 pKa = 11.84SSHH20 pKa = 6.49SYY22 pKa = 10.6DD23 pKa = 2.85IRR25 pKa = 11.84CCLFLCLSLWTEE37 pKa = 4.31FSQAGDD43 pKa = 3.56LEE45 pKa = 4.34YY46 pKa = 10.87LSRR49 pKa = 11.84GLKK52 pKa = 9.93DD53 pKa = 3.05VRR55 pKa = 11.84GFMTTDD61 pKa = 2.65NVYY64 pKa = 10.46INTTTRR70 pKa = 11.84VQDD73 pKa = 4.14FSEE76 pKa = 3.95SRR78 pKa = 11.84YY79 pKa = 9.81EE80 pKa = 4.0VLVHH84 pKa = 7.19NITEE88 pKa = 4.22KK89 pKa = 10.52MLTPTEE95 pKa = 3.94FMVYY99 pKa = 8.96DD100 pKa = 4.47TDD102 pKa = 5.39AGDD105 pKa = 4.1PFCTDD110 pKa = 2.96YY111 pKa = 10.74LTYY114 pKa = 10.85DD115 pKa = 3.44VGWFSKK121 pKa = 10.93NNPTKK126 pKa = 10.57EE127 pKa = 4.41CVNSLARR134 pKa = 11.84SNCFYY139 pKa = 10.78LWHH142 pKa = 6.32NHH144 pKa = 6.17DD145 pKa = 5.18DD146 pKa = 4.35YY147 pKa = 11.44EE148 pKa = 4.66ISWFEE153 pKa = 3.85NVHH156 pKa = 5.36TLVCVGIRR164 pKa = 11.84YY165 pKa = 9.53SKK167 pKa = 10.75SQITSRR173 pKa = 11.84KK174 pKa = 7.43NSHH177 pKa = 4.67WSRR180 pKa = 11.84WYY182 pKa = 10.2SLTRR186 pKa = 11.84AEE188 pKa = 4.37PTSILQYY195 pKa = 11.61GMIFKK200 pKa = 10.57NGEE203 pKa = 3.99YY204 pKa = 10.53HH205 pKa = 7.27LMSPMALEE213 pKa = 4.34FGLVYY218 pKa = 10.65DD219 pKa = 4.54LSRR222 pKa = 11.84NDD224 pKa = 3.64FYY226 pKa = 11.72GYY228 pKa = 8.94ATNITHH234 pKa = 7.56ALVLEE239 pKa = 4.04HH240 pKa = 7.19RR241 pKa = 11.84NLIIGSSPTLPAEE254 pKa = 4.72FKK256 pKa = 10.62HH257 pKa = 5.43ITYY260 pKa = 7.62TTLGKK265 pKa = 8.91KK266 pKa = 7.68TVFCNDD272 pKa = 4.05RR273 pKa = 11.84IVGNPIDD280 pKa = 5.26DD281 pKa = 4.22NEE283 pKa = 4.37SDD285 pKa = 3.86EE286 pKa = 5.62VIATDD291 pKa = 4.58FNCDD295 pKa = 2.99PHH297 pKa = 7.56QVLGVIQRR305 pKa = 11.84PNGDD309 pKa = 4.17LLPVYY314 pKa = 9.6WCLQWNCVNTKK325 pKa = 10.4SFGDD329 pKa = 3.83LFTTEE334 pKa = 4.44LNRR337 pKa = 11.84KK338 pKa = 9.44FEE340 pKa = 4.84DD341 pKa = 4.04DD342 pKa = 3.22EE343 pKa = 5.02HH344 pKa = 7.56EE345 pKa = 4.28NFAVRR350 pKa = 11.84ILLIIRR356 pKa = 11.84SLIVKK361 pKa = 8.08ATLYY365 pKa = 8.82ITQSINLRR373 pKa = 11.84VFSLSVWGIFTTIMVIRR390 pKa = 11.84LTGSWIAGTIMGLYY404 pKa = 10.01VVVLLSDD411 pKa = 3.79
MM1 pKa = 7.58GSYY4 pKa = 8.83WCSNFALLWVQKK16 pKa = 9.93RR17 pKa = 11.84SSHH20 pKa = 6.49SYY22 pKa = 10.6DD23 pKa = 2.85IRR25 pKa = 11.84CCLFLCLSLWTEE37 pKa = 4.31FSQAGDD43 pKa = 3.56LEE45 pKa = 4.34YY46 pKa = 10.87LSRR49 pKa = 11.84GLKK52 pKa = 9.93DD53 pKa = 3.05VRR55 pKa = 11.84GFMTTDD61 pKa = 2.65NVYY64 pKa = 10.46INTTTRR70 pKa = 11.84VQDD73 pKa = 4.14FSEE76 pKa = 3.95SRR78 pKa = 11.84YY79 pKa = 9.81EE80 pKa = 4.0VLVHH84 pKa = 7.19NITEE88 pKa = 4.22KK89 pKa = 10.52MLTPTEE95 pKa = 3.94FMVYY99 pKa = 8.96DD100 pKa = 4.47TDD102 pKa = 5.39AGDD105 pKa = 4.1PFCTDD110 pKa = 2.96YY111 pKa = 10.74LTYY114 pKa = 10.85DD115 pKa = 3.44VGWFSKK121 pKa = 10.93NNPTKK126 pKa = 10.57EE127 pKa = 4.41CVNSLARR134 pKa = 11.84SNCFYY139 pKa = 10.78LWHH142 pKa = 6.32NHH144 pKa = 6.17DD145 pKa = 5.18DD146 pKa = 4.35YY147 pKa = 11.44EE148 pKa = 4.66ISWFEE153 pKa = 3.85NVHH156 pKa = 5.36TLVCVGIRR164 pKa = 11.84YY165 pKa = 9.53SKK167 pKa = 10.75SQITSRR173 pKa = 11.84KK174 pKa = 7.43NSHH177 pKa = 4.67WSRR180 pKa = 11.84WYY182 pKa = 10.2SLTRR186 pKa = 11.84AEE188 pKa = 4.37PTSILQYY195 pKa = 11.61GMIFKK200 pKa = 10.57NGEE203 pKa = 3.99YY204 pKa = 10.53HH205 pKa = 7.27LMSPMALEE213 pKa = 4.34FGLVYY218 pKa = 10.65DD219 pKa = 4.54LSRR222 pKa = 11.84NDD224 pKa = 3.64FYY226 pKa = 11.72GYY228 pKa = 8.94ATNITHH234 pKa = 7.56ALVLEE239 pKa = 4.04HH240 pKa = 7.19RR241 pKa = 11.84NLIIGSSPTLPAEE254 pKa = 4.72FKK256 pKa = 10.62HH257 pKa = 5.43ITYY260 pKa = 7.62TTLGKK265 pKa = 8.91KK266 pKa = 7.68TVFCNDD272 pKa = 4.05RR273 pKa = 11.84IVGNPIDD280 pKa = 5.26DD281 pKa = 4.22NEE283 pKa = 4.37SDD285 pKa = 3.86EE286 pKa = 5.62VIATDD291 pKa = 4.58FNCDD295 pKa = 2.99PHH297 pKa = 7.56QVLGVIQRR305 pKa = 11.84PNGDD309 pKa = 4.17LLPVYY314 pKa = 9.6WCLQWNCVNTKK325 pKa = 10.4SFGDD329 pKa = 3.83LFTTEE334 pKa = 4.44LNRR337 pKa = 11.84KK338 pKa = 9.44FEE340 pKa = 4.84DD341 pKa = 4.04DD342 pKa = 3.22EE343 pKa = 5.02HH344 pKa = 7.56EE345 pKa = 4.28NFAVRR350 pKa = 11.84ILLIIRR356 pKa = 11.84SLIVKK361 pKa = 8.08ATLYY365 pKa = 8.82ITQSINLRR373 pKa = 11.84VFSLSVWGIFTTIMVIRR390 pKa = 11.84LTGSWIAGTIMGLYY404 pKa = 10.01VVVLLSDD411 pKa = 3.79
Molecular weight: 47.55 kDa
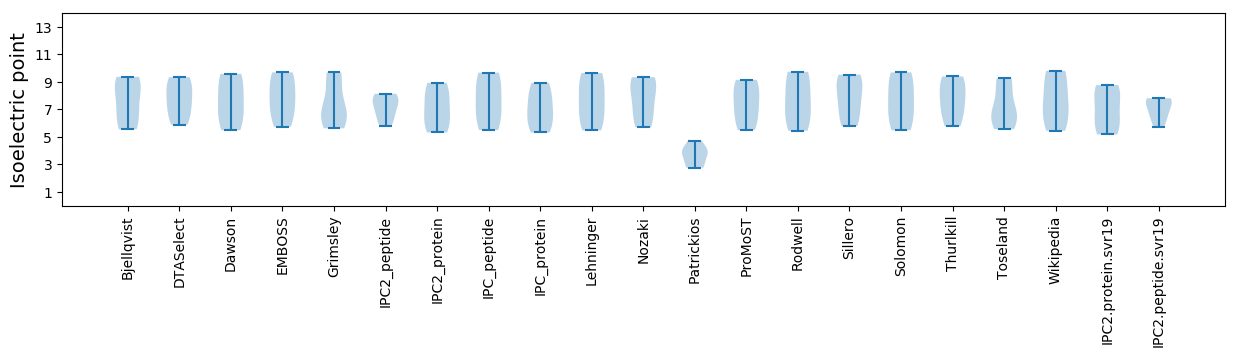
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0QKL9|A0A0P0QKL9_9VIRU Uncharacterized protein OS=Wuhan cricket virus OX=1746070 GN=VP3 PE=4 SV=1
MM1 pKa = 7.66NITKK5 pKa = 9.9VLLVALVGIHH15 pKa = 6.66FGGTAYY21 pKa = 10.56AATNTGASVAKK32 pKa = 9.29LTDD35 pKa = 2.65IWSIGSVSKK44 pKa = 10.48SWNPSDD50 pKa = 3.52WSDD53 pKa = 3.46DD54 pKa = 3.5FKK56 pKa = 11.66LRR58 pKa = 11.84VYY60 pKa = 10.73QSLTLVIAIAFLGGTMAHH78 pKa = 6.53MIVGFVVGIGLVHH91 pKa = 5.73NTWVLTGALILLYY104 pKa = 10.11YY105 pKa = 9.85GYY107 pKa = 11.0KK108 pKa = 9.65RR109 pKa = 11.84GHH111 pKa = 7.37PIAMILGAVCSYY123 pKa = 11.13VLAFGQSLAKK133 pKa = 10.61LGII136 pKa = 3.94
MM1 pKa = 7.66NITKK5 pKa = 9.9VLLVALVGIHH15 pKa = 6.66FGGTAYY21 pKa = 10.56AATNTGASVAKK32 pKa = 9.29LTDD35 pKa = 2.65IWSIGSVSKK44 pKa = 10.48SWNPSDD50 pKa = 3.52WSDD53 pKa = 3.46DD54 pKa = 3.5FKK56 pKa = 11.66LRR58 pKa = 11.84VYY60 pKa = 10.73QSLTLVIAIAFLGGTMAHH78 pKa = 6.53MIVGFVVGIGLVHH91 pKa = 5.73NTWVLTGALILLYY104 pKa = 10.11YY105 pKa = 9.85GYY107 pKa = 11.0KK108 pKa = 9.65RR109 pKa = 11.84GHH111 pKa = 7.37PIAMILGAVCSYY123 pKa = 11.13VLAFGQSLAKK133 pKa = 10.61LGII136 pKa = 3.94
Molecular weight: 14.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3094 |
136 |
943 |
515.7 |
58.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.527 ± 0.698 | 1.584 ± 0.321 |
4.622 ± 0.425 | 4.848 ± 0.62 |
3.943 ± 0.347 | 7.531 ± 0.736 |
2.618 ± 0.435 | 6.981 ± 0.438 |
4.654 ± 0.671 | 10.116 ± 0.318 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.846 ± 0.315 | 4.396 ± 0.363 |
4.525 ± 0.558 | 2.521 ± 0.199 |
5.139 ± 0.717 | 7.789 ± 1.051 |
6.27 ± 0.427 | 6.755 ± 0.458 |
1.875 ± 0.322 | 4.46 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
