
Capybara microvirus Cap1_SP_109
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
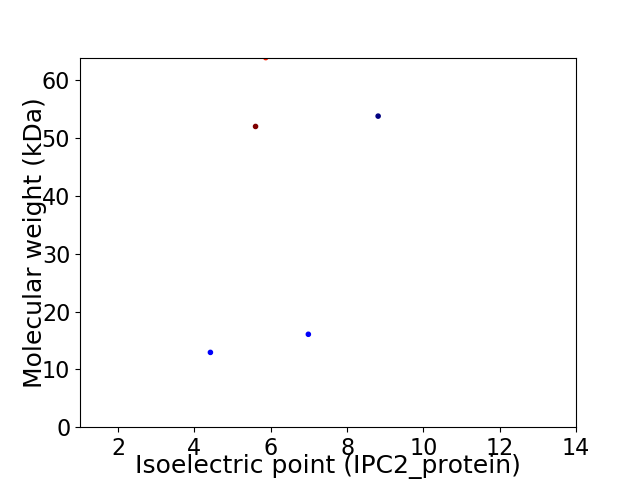
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
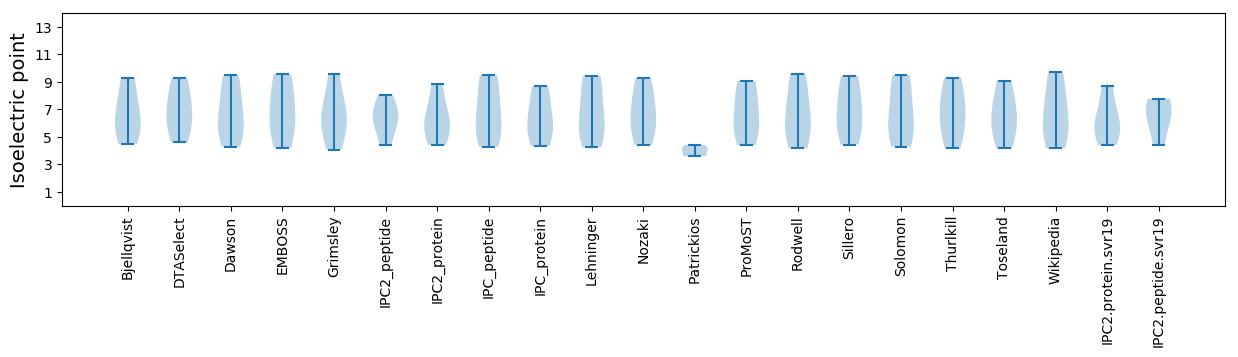
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W453|A0A4P8W453_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_109 OX=2584807 PE=4 SV=1
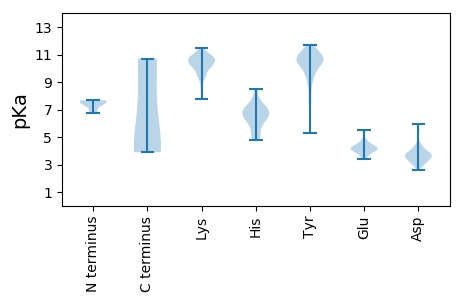
MM1 pKa = 7.68RR2 pKa = 11.84FYY4 pKa = 11.54VEE6 pKa = 4.44GSRR9 pKa = 11.84LFNRR13 pKa = 11.84STNEE17 pKa = 3.4DD18 pKa = 3.71SIKK21 pKa = 10.31VKK23 pKa = 10.67SYY25 pKa = 10.35VDD27 pKa = 3.74VIEE30 pKa = 4.31KK31 pKa = 10.09VVNEE35 pKa = 3.87KK36 pKa = 10.08TGLIEE41 pKa = 4.01EE42 pKa = 4.38KK43 pKa = 10.51VVCKK47 pKa = 10.37EE48 pKa = 4.07LTDD51 pKa = 3.71DD52 pKa = 4.05RR53 pKa = 11.84YY54 pKa = 10.95EE55 pKa = 4.34GLSSYY60 pKa = 11.35DD61 pKa = 3.14FDD63 pKa = 5.93LDD65 pKa = 4.27HH66 pKa = 7.07IVLAGATSLLQPCPKK81 pKa = 10.22VSGNVFDD88 pKa = 5.39ISDD91 pKa = 3.88SLASNVDD98 pKa = 3.42YY99 pKa = 11.4LGDD102 pKa = 3.61YY103 pKa = 11.35YY104 pKa = 10.58EE105 pKa = 4.23TNKK108 pKa = 10.47NIKK111 pKa = 9.6EE112 pKa = 4.1DD113 pKa = 3.63EE114 pKa = 4.13
MM1 pKa = 7.68RR2 pKa = 11.84FYY4 pKa = 11.54VEE6 pKa = 4.44GSRR9 pKa = 11.84LFNRR13 pKa = 11.84STNEE17 pKa = 3.4DD18 pKa = 3.71SIKK21 pKa = 10.31VKK23 pKa = 10.67SYY25 pKa = 10.35VDD27 pKa = 3.74VIEE30 pKa = 4.31KK31 pKa = 10.09VVNEE35 pKa = 3.87KK36 pKa = 10.08TGLIEE41 pKa = 4.01EE42 pKa = 4.38KK43 pKa = 10.51VVCKK47 pKa = 10.37EE48 pKa = 4.07LTDD51 pKa = 3.71DD52 pKa = 4.05RR53 pKa = 11.84YY54 pKa = 10.95EE55 pKa = 4.34GLSSYY60 pKa = 11.35DD61 pKa = 3.14FDD63 pKa = 5.93LDD65 pKa = 4.27HH66 pKa = 7.07IVLAGATSLLQPCPKK81 pKa = 10.22VSGNVFDD88 pKa = 5.39ISDD91 pKa = 3.88SLASNVDD98 pKa = 3.42YY99 pKa = 11.4LGDD102 pKa = 3.61YY103 pKa = 11.35YY104 pKa = 10.58EE105 pKa = 4.23TNKK108 pKa = 10.47NIKK111 pKa = 9.6EE112 pKa = 4.1DD113 pKa = 3.63EE114 pKa = 4.13
Molecular weight: 12.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Y4|A0A4P8W4Y4_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_109 OX=2584807 PE=4 SV=1
MM1 pKa = 7.57CLSPIKK7 pKa = 10.15IYY9 pKa = 10.65NRR11 pKa = 11.84SEE13 pKa = 4.1SLVTNGPLYY22 pKa = 10.59IEE24 pKa = 4.63VPCGKK29 pKa = 10.13CSEE32 pKa = 4.31CQKK35 pKa = 10.71QKK37 pKa = 9.31HH38 pKa = 4.92TDD40 pKa = 3.18YY41 pKa = 10.83CIRR44 pKa = 11.84AFWQYY49 pKa = 11.33QEE51 pKa = 4.13TLIKK55 pKa = 10.55GGYY58 pKa = 9.69CYY60 pKa = 10.59FDD62 pKa = 3.35TLTYY66 pKa = 10.71DD67 pKa = 3.45SSHH70 pKa = 6.4LPKK73 pKa = 10.19FDD75 pKa = 5.58GISCFDD81 pKa = 3.19KK82 pKa = 10.56THH84 pKa = 5.38IQKK87 pKa = 10.74FIRR90 pKa = 11.84DD91 pKa = 3.64LRR93 pKa = 11.84RR94 pKa = 11.84QLDD97 pKa = 3.45YY98 pKa = 11.29HH99 pKa = 6.7FYY101 pKa = 9.51EE102 pKa = 4.36SKK104 pKa = 10.76GRR106 pKa = 11.84SGSIKK111 pKa = 10.57EE112 pKa = 3.85NFKK115 pKa = 11.19YY116 pKa = 10.51FLTSEE121 pKa = 4.44YY122 pKa = 10.15GGKK125 pKa = 8.68RR126 pKa = 11.84HH127 pKa = 6.13RR128 pKa = 11.84PHH130 pKa = 6.21YY131 pKa = 10.12HH132 pKa = 4.96VLFFVTDD139 pKa = 3.75PSISVSQLKK148 pKa = 10.53YY149 pKa = 10.75YY150 pKa = 10.19INKK153 pKa = 8.55CWIYY157 pKa = 10.56GITDD161 pKa = 3.68SVSTTNDD168 pKa = 2.61RR169 pKa = 11.84VVRR172 pKa = 11.84GISAISYY179 pKa = 5.31VTKK182 pKa = 10.82YY183 pKa = 7.22VTKK186 pKa = 10.33DD187 pKa = 3.09QEE189 pKa = 4.05FSLLIEE195 pKa = 4.58KK196 pKa = 10.12RR197 pKa = 11.84LKK199 pKa = 10.66YY200 pKa = 10.79YY201 pKa = 10.71SILYY205 pKa = 10.43GSDD208 pKa = 2.77SLEE211 pKa = 3.88YY212 pKa = 10.56LEE214 pKa = 5.54KK215 pKa = 11.03VKK217 pKa = 10.44DD218 pKa = 3.81YY219 pKa = 11.4KK220 pKa = 10.63RR221 pKa = 11.84FRR223 pKa = 11.84PFFNSSTNFGSSALKK238 pKa = 10.13MVDD241 pKa = 3.65QDD243 pKa = 4.16LLLDD247 pKa = 3.65GRR249 pKa = 11.84IWISDD254 pKa = 3.36SKK256 pKa = 9.52TSKK259 pKa = 10.69KK260 pKa = 10.13FVSLPLYY267 pKa = 9.7YY268 pKa = 9.77RR269 pKa = 11.84RR270 pKa = 11.84KK271 pKa = 10.25LFYY274 pKa = 10.74DD275 pKa = 3.93VIKK278 pKa = 10.3TEE280 pKa = 4.34DD281 pKa = 3.65GRR283 pKa = 11.84IKK285 pKa = 10.27WVMNDD290 pKa = 2.75RR291 pKa = 11.84GFDD294 pKa = 3.68YY295 pKa = 10.68KK296 pKa = 10.92LKK298 pKa = 10.81RR299 pKa = 11.84LDD301 pKa = 3.66TNISLLEE308 pKa = 4.08DD309 pKa = 4.28RR310 pKa = 11.84YY311 pKa = 9.49NTYY314 pKa = 11.01YY315 pKa = 11.33LNLDD319 pKa = 3.76NYY321 pKa = 10.44CSEE324 pKa = 5.03DD325 pKa = 4.25YY326 pKa = 10.74FPKK329 pKa = 10.42DD330 pKa = 3.3YY331 pKa = 10.92VSRR334 pKa = 11.84LILSYY339 pKa = 11.34LDD341 pKa = 3.16GRR343 pKa = 11.84TFKK346 pKa = 11.09DD347 pKa = 3.4FATYY351 pKa = 10.25LYY353 pKa = 10.3VYY355 pKa = 8.95RR356 pKa = 11.84GKK358 pKa = 9.53MFSGGVLPDD367 pKa = 3.06FHH369 pKa = 8.48KK370 pKa = 10.57FYY372 pKa = 11.39SMSLRR377 pKa = 11.84GSTIFSKK384 pKa = 10.68LYY386 pKa = 10.43SSNLSIRR393 pKa = 11.84SRR395 pKa = 11.84MRR397 pKa = 11.84SRR399 pKa = 11.84LDD401 pKa = 3.4KK402 pKa = 10.79ISIRR406 pKa = 11.84EE407 pKa = 3.83TSFSSFRR414 pKa = 11.84NFDD417 pKa = 3.12KK418 pKa = 10.91LYY420 pKa = 10.84NLFNVLLRR428 pKa = 11.84EE429 pKa = 4.29SHH431 pKa = 6.69TSNQYY436 pKa = 8.9TFDD439 pKa = 3.85YY440 pKa = 11.05KK441 pKa = 11.02EE442 pKa = 4.17DD443 pKa = 4.52LIGKK447 pKa = 9.14YY448 pKa = 10.35KK449 pKa = 10.97LILL452 pKa = 3.89
MM1 pKa = 7.57CLSPIKK7 pKa = 10.15IYY9 pKa = 10.65NRR11 pKa = 11.84SEE13 pKa = 4.1SLVTNGPLYY22 pKa = 10.59IEE24 pKa = 4.63VPCGKK29 pKa = 10.13CSEE32 pKa = 4.31CQKK35 pKa = 10.71QKK37 pKa = 9.31HH38 pKa = 4.92TDD40 pKa = 3.18YY41 pKa = 10.83CIRR44 pKa = 11.84AFWQYY49 pKa = 11.33QEE51 pKa = 4.13TLIKK55 pKa = 10.55GGYY58 pKa = 9.69CYY60 pKa = 10.59FDD62 pKa = 3.35TLTYY66 pKa = 10.71DD67 pKa = 3.45SSHH70 pKa = 6.4LPKK73 pKa = 10.19FDD75 pKa = 5.58GISCFDD81 pKa = 3.19KK82 pKa = 10.56THH84 pKa = 5.38IQKK87 pKa = 10.74FIRR90 pKa = 11.84DD91 pKa = 3.64LRR93 pKa = 11.84RR94 pKa = 11.84QLDD97 pKa = 3.45YY98 pKa = 11.29HH99 pKa = 6.7FYY101 pKa = 9.51EE102 pKa = 4.36SKK104 pKa = 10.76GRR106 pKa = 11.84SGSIKK111 pKa = 10.57EE112 pKa = 3.85NFKK115 pKa = 11.19YY116 pKa = 10.51FLTSEE121 pKa = 4.44YY122 pKa = 10.15GGKK125 pKa = 8.68RR126 pKa = 11.84HH127 pKa = 6.13RR128 pKa = 11.84PHH130 pKa = 6.21YY131 pKa = 10.12HH132 pKa = 4.96VLFFVTDD139 pKa = 3.75PSISVSQLKK148 pKa = 10.53YY149 pKa = 10.75YY150 pKa = 10.19INKK153 pKa = 8.55CWIYY157 pKa = 10.56GITDD161 pKa = 3.68SVSTTNDD168 pKa = 2.61RR169 pKa = 11.84VVRR172 pKa = 11.84GISAISYY179 pKa = 5.31VTKK182 pKa = 10.82YY183 pKa = 7.22VTKK186 pKa = 10.33DD187 pKa = 3.09QEE189 pKa = 4.05FSLLIEE195 pKa = 4.58KK196 pKa = 10.12RR197 pKa = 11.84LKK199 pKa = 10.66YY200 pKa = 10.79YY201 pKa = 10.71SILYY205 pKa = 10.43GSDD208 pKa = 2.77SLEE211 pKa = 3.88YY212 pKa = 10.56LEE214 pKa = 5.54KK215 pKa = 11.03VKK217 pKa = 10.44DD218 pKa = 3.81YY219 pKa = 11.4KK220 pKa = 10.63RR221 pKa = 11.84FRR223 pKa = 11.84PFFNSSTNFGSSALKK238 pKa = 10.13MVDD241 pKa = 3.65QDD243 pKa = 4.16LLLDD247 pKa = 3.65GRR249 pKa = 11.84IWISDD254 pKa = 3.36SKK256 pKa = 9.52TSKK259 pKa = 10.69KK260 pKa = 10.13FVSLPLYY267 pKa = 9.7YY268 pKa = 9.77RR269 pKa = 11.84RR270 pKa = 11.84KK271 pKa = 10.25LFYY274 pKa = 10.74DD275 pKa = 3.93VIKK278 pKa = 10.3TEE280 pKa = 4.34DD281 pKa = 3.65GRR283 pKa = 11.84IKK285 pKa = 10.27WVMNDD290 pKa = 2.75RR291 pKa = 11.84GFDD294 pKa = 3.68YY295 pKa = 10.68KK296 pKa = 10.92LKK298 pKa = 10.81RR299 pKa = 11.84LDD301 pKa = 3.66TNISLLEE308 pKa = 4.08DD309 pKa = 4.28RR310 pKa = 11.84YY311 pKa = 9.49NTYY314 pKa = 11.01YY315 pKa = 11.33LNLDD319 pKa = 3.76NYY321 pKa = 10.44CSEE324 pKa = 5.03DD325 pKa = 4.25YY326 pKa = 10.74FPKK329 pKa = 10.42DD330 pKa = 3.3YY331 pKa = 10.92VSRR334 pKa = 11.84LILSYY339 pKa = 11.34LDD341 pKa = 3.16GRR343 pKa = 11.84TFKK346 pKa = 11.09DD347 pKa = 3.4FATYY351 pKa = 10.25LYY353 pKa = 10.3VYY355 pKa = 8.95RR356 pKa = 11.84GKK358 pKa = 9.53MFSGGVLPDD367 pKa = 3.06FHH369 pKa = 8.48KK370 pKa = 10.57FYY372 pKa = 11.39SMSLRR377 pKa = 11.84GSTIFSKK384 pKa = 10.68LYY386 pKa = 10.43SSNLSIRR393 pKa = 11.84SRR395 pKa = 11.84MRR397 pKa = 11.84SRR399 pKa = 11.84LDD401 pKa = 3.4KK402 pKa = 10.79ISIRR406 pKa = 11.84EE407 pKa = 3.83TSFSSFRR414 pKa = 11.84NFDD417 pKa = 3.12KK418 pKa = 10.91LYY420 pKa = 10.84NLFNVLLRR428 pKa = 11.84EE429 pKa = 4.29SHH431 pKa = 6.69TSNQYY436 pKa = 8.9TFDD439 pKa = 3.85YY440 pKa = 11.05KK441 pKa = 11.02EE442 pKa = 4.17DD443 pKa = 4.52LIGKK447 pKa = 9.14YY448 pKa = 10.35KK449 pKa = 10.97LILL452 pKa = 3.89
Molecular weight: 53.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1743 |
114 |
570 |
348.6 |
39.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.475 ± 1.436 | 0.975 ± 0.361 |
6.942 ± 0.562 | 5.049 ± 0.717 |
4.877 ± 0.625 | 6.368 ± 0.855 |
1.205 ± 0.237 | 5.393 ± 0.389 |
6.942 ± 0.914 | 9.122 ± 1.145 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.721 ± 0.335 | 5.967 ± 1.06 |
2.582 ± 0.614 | 3.442 ± 0.954 |
5.278 ± 0.531 | 9.237 ± 0.588 |
6.311 ± 0.435 | 6.713 ± 0.916 |
0.688 ± 0.128 | 6.713 ± 1.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
