
Larimichthys crocea (Large yellow croaker) (Pseudosciaena crocea)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala;
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
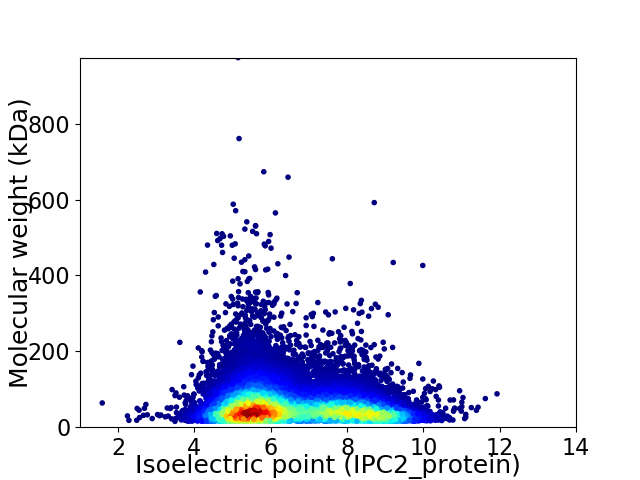
Virtual 2D-PAGE plot for 23960 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G0HV70|A0A6G0HV70_LARCR Adenylosuccinate lyase OS=Larimichthys crocea OX=215358 GN=D5F01_LYC18357 PE=3 SV=1
MM1 pKa = 7.58PAVNLIKK8 pKa = 10.31VTTNPAQTDD17 pKa = 3.14DD18 pKa = 3.64SQRR21 pKa = 11.84YY22 pKa = 6.68KK23 pKa = 10.77TDD25 pKa = 3.17VNPLTRR31 pKa = 11.84PSGLDD36 pKa = 3.17NLIEE40 pKa = 4.41EE41 pKa = 4.36DD42 pKa = 4.13TEE44 pKa = 4.17PKK46 pKa = 10.16EE47 pKa = 3.9EE48 pKa = 4.06VMGVFSSYY56 pKa = 11.01NGNPADD62 pKa = 3.35QYY64 pKa = 11.79ARR66 pKa = 11.84VSKK69 pKa = 10.67ISVDD73 pKa = 3.65SFDD76 pKa = 4.22SSSTSSGEE84 pKa = 4.2CYY86 pKa = 10.4EE87 pKa = 4.08NTSNYY92 pKa = 7.03VTPEE96 pKa = 4.13PGPSQSYY103 pKa = 8.09TVNDD107 pKa = 4.25PGDD110 pKa = 3.67FPSGYY115 pKa = 10.07RR116 pKa = 11.84NNQQTTDD123 pKa = 3.62QHH125 pKa = 5.72WLNPAEE131 pKa = 4.28VDD133 pKa = 3.54DD134 pKa = 5.69GIYY137 pKa = 10.72SPVSPDD143 pKa = 3.39QDD145 pKa = 3.4PSSDD149 pKa = 4.72DD150 pKa = 4.36DD151 pKa = 4.65YY152 pKa = 12.05DD153 pKa = 6.25DD154 pKa = 4.41IGAA157 pKa = 4.41
MM1 pKa = 7.58PAVNLIKK8 pKa = 10.31VTTNPAQTDD17 pKa = 3.14DD18 pKa = 3.64SQRR21 pKa = 11.84YY22 pKa = 6.68KK23 pKa = 10.77TDD25 pKa = 3.17VNPLTRR31 pKa = 11.84PSGLDD36 pKa = 3.17NLIEE40 pKa = 4.41EE41 pKa = 4.36DD42 pKa = 4.13TEE44 pKa = 4.17PKK46 pKa = 10.16EE47 pKa = 3.9EE48 pKa = 4.06VMGVFSSYY56 pKa = 11.01NGNPADD62 pKa = 3.35QYY64 pKa = 11.79ARR66 pKa = 11.84VSKK69 pKa = 10.67ISVDD73 pKa = 3.65SFDD76 pKa = 4.22SSSTSSGEE84 pKa = 4.2CYY86 pKa = 10.4EE87 pKa = 4.08NTSNYY92 pKa = 7.03VTPEE96 pKa = 4.13PGPSQSYY103 pKa = 8.09TVNDD107 pKa = 4.25PGDD110 pKa = 3.67FPSGYY115 pKa = 10.07RR116 pKa = 11.84NNQQTTDD123 pKa = 3.62QHH125 pKa = 5.72WLNPAEE131 pKa = 4.28VDD133 pKa = 3.54DD134 pKa = 5.69GIYY137 pKa = 10.72SPVSPDD143 pKa = 3.39QDD145 pKa = 3.4PSSDD149 pKa = 4.72DD150 pKa = 4.36DD151 pKa = 4.65YY152 pKa = 12.05DD153 pKa = 6.25DD154 pKa = 4.41IGAA157 pKa = 4.41
Molecular weight: 17.19 kDa
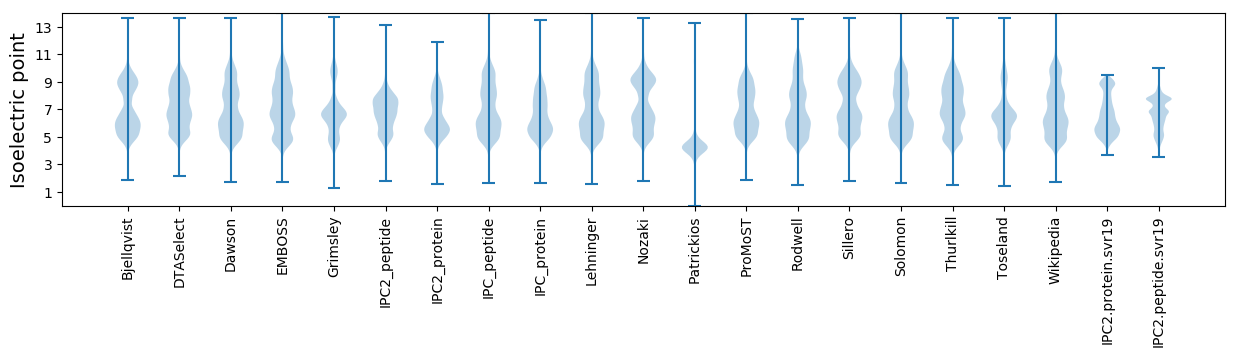
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G0J7C5|A0A6G0J7C5_LARCR RNA helicase OS=Larimichthys crocea OX=215358 GN=D5F01_LYC01755 PE=3 SV=1
MM1 pKa = 7.61LLLGVHH7 pKa = 6.47TKK9 pKa = 10.49RR10 pKa = 11.84KK11 pKa = 6.46TLRR14 pKa = 11.84LALTNRR20 pKa = 11.84ITNRR24 pKa = 11.84NINRR28 pKa = 11.84TINSNINKK36 pKa = 6.18TTNRR40 pKa = 11.84TLNKK44 pKa = 6.38TTNRR48 pKa = 11.84TINKK52 pKa = 5.6TTNRR56 pKa = 11.84TINKK60 pKa = 6.0TTNKK64 pKa = 10.01NINRR68 pKa = 11.84TITKK72 pKa = 5.86TTNRR76 pKa = 11.84TINKK80 pKa = 7.91TINKK84 pKa = 6.12TTNKK88 pKa = 9.26TINKK92 pKa = 5.65TTNRR96 pKa = 11.84TINKK100 pKa = 5.52TTNRR104 pKa = 11.84NIKK107 pKa = 5.99TTNRR111 pKa = 11.84TTNRR115 pKa = 11.84TTNRR119 pKa = 11.84TINKK123 pKa = 5.52TTNRR127 pKa = 11.84NIKK130 pKa = 6.05TTNRR134 pKa = 11.84NIKK137 pKa = 6.12TTNRR141 pKa = 11.84TINKK145 pKa = 5.52TTNRR149 pKa = 11.84NIKK152 pKa = 9.41NHH154 pKa = 4.75QQKK157 pKa = 9.24HH158 pKa = 4.32QQNHH162 pKa = 5.15HH163 pKa = 5.78QNHH166 pKa = 4.89QQNHH170 pKa = 4.3QQNHH174 pKa = 4.39QQNHH178 pKa = 4.23QQNHH182 pKa = 4.82QEE184 pKa = 3.87NDD186 pKa = 3.61QQKK189 pKa = 9.0HH190 pKa = 4.4QQNHH194 pKa = 5.15HH195 pKa = 5.78QNHH198 pKa = 4.89QQNHH202 pKa = 4.3QQNHH206 pKa = 4.12QQNYY210 pKa = 5.49QQNDD214 pKa = 3.65QQKK217 pKa = 8.47HH218 pKa = 4.19QQNHH222 pKa = 5.15HH223 pKa = 5.78QNHH226 pKa = 4.89QQNHH230 pKa = 4.3QQNHH234 pKa = 4.04QQNINKK240 pKa = 6.62TTNRR244 pKa = 11.84TINKK248 pKa = 5.6TTNRR252 pKa = 11.84TINKK256 pKa = 5.51TTNRR260 pKa = 11.84TTNRR264 pKa = 11.84TINKK268 pKa = 6.14TTNKK272 pKa = 9.26TINKK276 pKa = 5.65TTNRR280 pKa = 11.84TITKK284 pKa = 5.29TTNRR288 pKa = 11.84TTNRR292 pKa = 11.84TINKK296 pKa = 6.14TTNKK300 pKa = 9.26TINKK304 pKa = 5.55TTNRR308 pKa = 11.84NIKK311 pKa = 5.99TTNRR315 pKa = 11.84TTNRR319 pKa = 11.84TINKK323 pKa = 5.52TTNRR327 pKa = 11.84NIKK330 pKa = 6.12TTNRR334 pKa = 11.84TINKK338 pKa = 5.52TTNRR342 pKa = 11.84NIKK345 pKa = 5.99TTNRR349 pKa = 11.84TTNRR353 pKa = 11.84TINKK357 pKa = 5.52TTNRR361 pKa = 11.84NIKK364 pKa = 6.12TTNRR368 pKa = 11.84TINKK372 pKa = 5.52TTNRR376 pKa = 11.84NIKK379 pKa = 5.99TTNRR383 pKa = 11.84TTNRR387 pKa = 11.84TINKK391 pKa = 5.51TTNRR395 pKa = 11.84TTNRR399 pKa = 11.84TINKK403 pKa = 5.52TTNRR407 pKa = 11.84NIKK410 pKa = 6.07TTNRR414 pKa = 11.84NINRR418 pKa = 11.84TINRR422 pKa = 11.84TINRR426 pKa = 11.84TTNKK430 pKa = 5.61TTNRR434 pKa = 11.84KK435 pKa = 8.92
MM1 pKa = 7.61LLLGVHH7 pKa = 6.47TKK9 pKa = 10.49RR10 pKa = 11.84KK11 pKa = 6.46TLRR14 pKa = 11.84LALTNRR20 pKa = 11.84ITNRR24 pKa = 11.84NINRR28 pKa = 11.84TINSNINKK36 pKa = 6.18TTNRR40 pKa = 11.84TLNKK44 pKa = 6.38TTNRR48 pKa = 11.84TINKK52 pKa = 5.6TTNRR56 pKa = 11.84TINKK60 pKa = 6.0TTNKK64 pKa = 10.01NINRR68 pKa = 11.84TITKK72 pKa = 5.86TTNRR76 pKa = 11.84TINKK80 pKa = 7.91TINKK84 pKa = 6.12TTNKK88 pKa = 9.26TINKK92 pKa = 5.65TTNRR96 pKa = 11.84TINKK100 pKa = 5.52TTNRR104 pKa = 11.84NIKK107 pKa = 5.99TTNRR111 pKa = 11.84TTNRR115 pKa = 11.84TTNRR119 pKa = 11.84TINKK123 pKa = 5.52TTNRR127 pKa = 11.84NIKK130 pKa = 6.05TTNRR134 pKa = 11.84NIKK137 pKa = 6.12TTNRR141 pKa = 11.84TINKK145 pKa = 5.52TTNRR149 pKa = 11.84NIKK152 pKa = 9.41NHH154 pKa = 4.75QQKK157 pKa = 9.24HH158 pKa = 4.32QQNHH162 pKa = 5.15HH163 pKa = 5.78QNHH166 pKa = 4.89QQNHH170 pKa = 4.3QQNHH174 pKa = 4.39QQNHH178 pKa = 4.23QQNHH182 pKa = 4.82QEE184 pKa = 3.87NDD186 pKa = 3.61QQKK189 pKa = 9.0HH190 pKa = 4.4QQNHH194 pKa = 5.15HH195 pKa = 5.78QNHH198 pKa = 4.89QQNHH202 pKa = 4.3QQNHH206 pKa = 4.12QQNYY210 pKa = 5.49QQNDD214 pKa = 3.65QQKK217 pKa = 8.47HH218 pKa = 4.19QQNHH222 pKa = 5.15HH223 pKa = 5.78QNHH226 pKa = 4.89QQNHH230 pKa = 4.3QQNHH234 pKa = 4.04QQNINKK240 pKa = 6.62TTNRR244 pKa = 11.84TINKK248 pKa = 5.6TTNRR252 pKa = 11.84TINKK256 pKa = 5.51TTNRR260 pKa = 11.84TTNRR264 pKa = 11.84TINKK268 pKa = 6.14TTNKK272 pKa = 9.26TINKK276 pKa = 5.65TTNRR280 pKa = 11.84TITKK284 pKa = 5.29TTNRR288 pKa = 11.84TTNRR292 pKa = 11.84TINKK296 pKa = 6.14TTNKK300 pKa = 9.26TINKK304 pKa = 5.55TTNRR308 pKa = 11.84NIKK311 pKa = 5.99TTNRR315 pKa = 11.84TTNRR319 pKa = 11.84TINKK323 pKa = 5.52TTNRR327 pKa = 11.84NIKK330 pKa = 6.12TTNRR334 pKa = 11.84TINKK338 pKa = 5.52TTNRR342 pKa = 11.84NIKK345 pKa = 5.99TTNRR349 pKa = 11.84TTNRR353 pKa = 11.84TINKK357 pKa = 5.52TTNRR361 pKa = 11.84NIKK364 pKa = 6.12TTNRR368 pKa = 11.84TINKK372 pKa = 5.52TTNRR376 pKa = 11.84NIKK379 pKa = 5.99TTNRR383 pKa = 11.84TTNRR387 pKa = 11.84TINKK391 pKa = 5.51TTNRR395 pKa = 11.84TTNRR399 pKa = 11.84TINKK403 pKa = 5.52TTNRR407 pKa = 11.84NIKK410 pKa = 6.07TTNRR414 pKa = 11.84NINRR418 pKa = 11.84TINRR422 pKa = 11.84TINRR426 pKa = 11.84TTNKK430 pKa = 5.61TTNRR434 pKa = 11.84KK435 pKa = 8.92
Molecular weight: 51.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13034180 |
150 |
16976 |
544.0 |
60.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.594 ± 0.015 | 2.198 ± 0.013 |
5.244 ± 0.012 | 6.933 ± 0.023 |
3.425 ± 0.012 | 6.327 ± 0.022 |
2.717 ± 0.009 | 4.228 ± 0.01 |
5.547 ± 0.017 | 9.38 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.008 | 3.816 ± 0.013 |
5.847 ± 0.02 | 4.823 ± 0.015 |
5.795 ± 0.016 | 8.724 ± 0.021 |
5.84 ± 0.014 | 6.331 ± 0.015 |
1.158 ± 0.005 | 2.668 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
