
Haloactinopolyspora alba
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Haloactinopolyspora
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
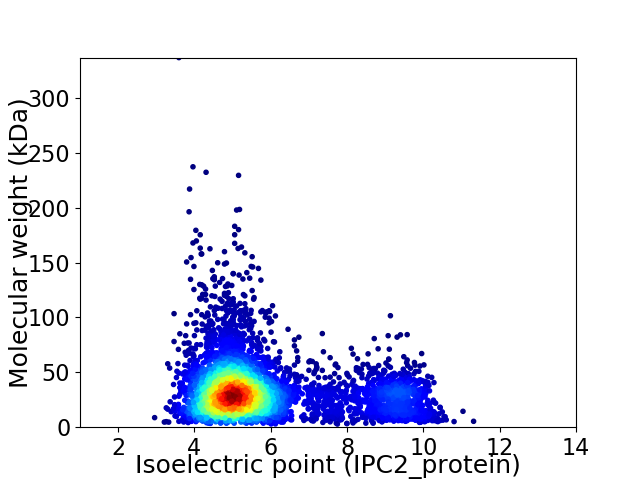
Virtual 2D-PAGE plot for 5159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P8DX69|A0A2P8DX69_9ACTN Carbohydrate ABC transporter membrane protein 2 (CUT1 family) OS=Haloactinopolyspora alba OX=648780 GN=CLV30_1126 PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84PSRR5 pKa = 11.84SRR7 pKa = 11.84SSAAIGASVTALAMIAVAIPAQADD31 pKa = 3.46DD32 pKa = 4.8HH33 pKa = 6.78EE34 pKa = 4.8NSPGEE39 pKa = 4.0PTITLPINDD48 pKa = 3.78SAAGGTYY55 pKa = 10.54DD56 pKa = 4.24QDD58 pKa = 3.27FTRR61 pKa = 11.84AYY63 pKa = 10.41AGEE66 pKa = 4.42TPDD69 pKa = 3.43GTPVYY74 pKa = 10.29VYY76 pKa = 10.69VPAGTPLDD84 pKa = 3.91GSAPAGVPSLDD95 pKa = 3.39PQFDD99 pKa = 4.18HH100 pKa = 7.14NPDD103 pKa = 3.88DD104 pKa = 4.01EE105 pKa = 4.71FVPCEE110 pKa = 3.88TATAGQFTLTQAQIDD125 pKa = 4.21YY126 pKa = 10.16VGDD129 pKa = 3.58MLAEE133 pKa = 4.45RR134 pKa = 11.84IVAVDD139 pKa = 3.71EE140 pKa = 4.3EE141 pKa = 5.11HH142 pKa = 6.88FGAMDD147 pKa = 4.0AADD150 pKa = 4.4PSDD153 pKa = 3.96PASDD157 pKa = 3.63SLVMLVYY164 pKa = 10.46NVQDD168 pKa = 3.44DD169 pKa = 4.52AYY171 pKa = 9.46YY172 pKa = 10.85DD173 pKa = 3.91CAATSYY179 pKa = 9.15TAGYY183 pKa = 7.64FAPDD187 pKa = 5.03FIDD190 pKa = 3.43SSGMNTIVVDD200 pKa = 4.01GLDD203 pKa = 3.01WANRR207 pKa = 11.84VGPNDD212 pKa = 3.68AEE214 pKa = 4.23WRR216 pKa = 11.84DD217 pKa = 3.47GDD219 pKa = 4.03PANDD223 pKa = 3.59RR224 pKa = 11.84PEE226 pKa = 4.58LYY228 pKa = 10.04EE229 pKa = 3.88GVVAHH234 pKa = 6.13EE235 pKa = 4.75LEE237 pKa = 4.83HH238 pKa = 6.9LLHH241 pKa = 6.75NYY243 pKa = 10.07GDD245 pKa = 4.06GGEE248 pKa = 4.34LSWVDD253 pKa = 3.59EE254 pKa = 4.31GLADD258 pKa = 5.01LAVFLNGFDD267 pKa = 3.79AGGSHH272 pKa = 7.53LSNHH276 pKa = 4.28QVFYY280 pKa = 11.23EE281 pKa = 4.29EE282 pKa = 4.26TSLTRR287 pKa = 11.84WGGSLANYY295 pKa = 8.39GAAFTYY301 pKa = 9.16FQYY304 pKa = 10.98LWEE307 pKa = 4.13QAGGNGDD314 pKa = 3.77GTFTPDD320 pKa = 3.08QQYY323 pKa = 11.09DD324 pKa = 3.39AAAGDD329 pKa = 4.37LLIKK333 pKa = 9.85TIFEE337 pKa = 4.01EE338 pKa = 4.47QANGMAGVQNAIDD351 pKa = 3.88TFNAATGSDD360 pKa = 3.7LRR362 pKa = 11.84SAEE365 pKa = 4.35ALFKK369 pKa = 10.22DD370 pKa = 3.56WSVAVYY376 pKa = 10.67LDD378 pKa = 4.72DD379 pKa = 4.34EE380 pKa = 4.34QSPVYY385 pKa = 9.98DD386 pKa = 3.55IKK388 pKa = 11.4AFDD391 pKa = 4.14FGDD394 pKa = 4.21PEE396 pKa = 4.29DD397 pKa = 4.63TSWTMDD403 pKa = 3.22IADD406 pKa = 3.55DD407 pKa = 4.21TFWGGRR413 pKa = 11.84GSYY416 pKa = 9.7QGSQPEE422 pKa = 4.46AKK424 pKa = 9.48WSRR427 pKa = 11.84LKK429 pKa = 10.71NRR431 pKa = 11.84PDD433 pKa = 3.07TTAVPFGMSVEE444 pKa = 4.27RR445 pKa = 11.84FRR447 pKa = 11.84NPGPSVRR454 pKa = 11.84VDD456 pKa = 3.74FDD458 pKa = 4.33GDD460 pKa = 3.55DD461 pKa = 3.63RR462 pKa = 11.84TQVAPHH468 pKa = 6.8AGDD471 pKa = 3.47THH473 pKa = 6.8WYY475 pKa = 9.64GGYY478 pKa = 10.11EE479 pKa = 4.07SQADD483 pKa = 3.45NVLNVDD489 pKa = 4.02AGTSPSTLDD498 pKa = 3.16FWSWNFIEE506 pKa = 4.85QGWDD510 pKa = 3.21YY511 pKa = 11.43GFVEE515 pKa = 4.48ALVGGEE521 pKa = 4.07WVTVPLVDD529 pKa = 5.5DD530 pKa = 4.52EE531 pKa = 5.38GNTVTTDD538 pKa = 4.28DD539 pKa = 5.04DD540 pKa = 3.83PHH542 pKa = 7.9GNNTEE547 pKa = 4.32GNGLTGTSGGEE558 pKa = 4.02YY559 pKa = 10.35FVDD562 pKa = 3.79DD563 pKa = 4.16PVYY566 pKa = 11.09VHH568 pKa = 6.72LTAQLPAGATDD579 pKa = 2.98VRR581 pKa = 11.84FRR583 pKa = 11.84YY584 pKa = 8.58STDD587 pKa = 2.83AAYY590 pKa = 10.92LDD592 pKa = 4.21TGWFVDD598 pKa = 3.97DD599 pKa = 3.96VRR601 pKa = 11.84VDD603 pKa = 3.47GAEE606 pKa = 3.9ASVDD610 pKa = 3.19SDD612 pKa = 3.28AGHH615 pKa = 6.24WTRR618 pKa = 11.84TTGVQEE624 pKa = 4.32NNWTVQTIAACDD636 pKa = 3.85LTPGTTLPGEE646 pKa = 4.47SSDD649 pKa = 3.6GAGNHH654 pKa = 5.53VYY656 pKa = 10.39RR657 pKa = 11.84FEE659 pKa = 6.17GDD661 pKa = 3.1AFEE664 pKa = 4.67QGGLEE669 pKa = 4.23TKK671 pKa = 10.18CANGNQDD678 pKa = 4.15DD679 pKa = 4.25FAVTISNLPTGDD691 pKa = 3.92LTFLDD696 pKa = 3.43ADD698 pKa = 3.79YY699 pKa = 10.31TFRR702 pKa = 11.84VANTGNTKK710 pKa = 10.52
MM1 pKa = 7.58RR2 pKa = 11.84PSRR5 pKa = 11.84SRR7 pKa = 11.84SSAAIGASVTALAMIAVAIPAQADD31 pKa = 3.46DD32 pKa = 4.8HH33 pKa = 6.78EE34 pKa = 4.8NSPGEE39 pKa = 4.0PTITLPINDD48 pKa = 3.78SAAGGTYY55 pKa = 10.54DD56 pKa = 4.24QDD58 pKa = 3.27FTRR61 pKa = 11.84AYY63 pKa = 10.41AGEE66 pKa = 4.42TPDD69 pKa = 3.43GTPVYY74 pKa = 10.29VYY76 pKa = 10.69VPAGTPLDD84 pKa = 3.91GSAPAGVPSLDD95 pKa = 3.39PQFDD99 pKa = 4.18HH100 pKa = 7.14NPDD103 pKa = 3.88DD104 pKa = 4.01EE105 pKa = 4.71FVPCEE110 pKa = 3.88TATAGQFTLTQAQIDD125 pKa = 4.21YY126 pKa = 10.16VGDD129 pKa = 3.58MLAEE133 pKa = 4.45RR134 pKa = 11.84IVAVDD139 pKa = 3.71EE140 pKa = 4.3EE141 pKa = 5.11HH142 pKa = 6.88FGAMDD147 pKa = 4.0AADD150 pKa = 4.4PSDD153 pKa = 3.96PASDD157 pKa = 3.63SLVMLVYY164 pKa = 10.46NVQDD168 pKa = 3.44DD169 pKa = 4.52AYY171 pKa = 9.46YY172 pKa = 10.85DD173 pKa = 3.91CAATSYY179 pKa = 9.15TAGYY183 pKa = 7.64FAPDD187 pKa = 5.03FIDD190 pKa = 3.43SSGMNTIVVDD200 pKa = 4.01GLDD203 pKa = 3.01WANRR207 pKa = 11.84VGPNDD212 pKa = 3.68AEE214 pKa = 4.23WRR216 pKa = 11.84DD217 pKa = 3.47GDD219 pKa = 4.03PANDD223 pKa = 3.59RR224 pKa = 11.84PEE226 pKa = 4.58LYY228 pKa = 10.04EE229 pKa = 3.88GVVAHH234 pKa = 6.13EE235 pKa = 4.75LEE237 pKa = 4.83HH238 pKa = 6.9LLHH241 pKa = 6.75NYY243 pKa = 10.07GDD245 pKa = 4.06GGEE248 pKa = 4.34LSWVDD253 pKa = 3.59EE254 pKa = 4.31GLADD258 pKa = 5.01LAVFLNGFDD267 pKa = 3.79AGGSHH272 pKa = 7.53LSNHH276 pKa = 4.28QVFYY280 pKa = 11.23EE281 pKa = 4.29EE282 pKa = 4.26TSLTRR287 pKa = 11.84WGGSLANYY295 pKa = 8.39GAAFTYY301 pKa = 9.16FQYY304 pKa = 10.98LWEE307 pKa = 4.13QAGGNGDD314 pKa = 3.77GTFTPDD320 pKa = 3.08QQYY323 pKa = 11.09DD324 pKa = 3.39AAAGDD329 pKa = 4.37LLIKK333 pKa = 9.85TIFEE337 pKa = 4.01EE338 pKa = 4.47QANGMAGVQNAIDD351 pKa = 3.88TFNAATGSDD360 pKa = 3.7LRR362 pKa = 11.84SAEE365 pKa = 4.35ALFKK369 pKa = 10.22DD370 pKa = 3.56WSVAVYY376 pKa = 10.67LDD378 pKa = 4.72DD379 pKa = 4.34EE380 pKa = 4.34QSPVYY385 pKa = 9.98DD386 pKa = 3.55IKK388 pKa = 11.4AFDD391 pKa = 4.14FGDD394 pKa = 4.21PEE396 pKa = 4.29DD397 pKa = 4.63TSWTMDD403 pKa = 3.22IADD406 pKa = 3.55DD407 pKa = 4.21TFWGGRR413 pKa = 11.84GSYY416 pKa = 9.7QGSQPEE422 pKa = 4.46AKK424 pKa = 9.48WSRR427 pKa = 11.84LKK429 pKa = 10.71NRR431 pKa = 11.84PDD433 pKa = 3.07TTAVPFGMSVEE444 pKa = 4.27RR445 pKa = 11.84FRR447 pKa = 11.84NPGPSVRR454 pKa = 11.84VDD456 pKa = 3.74FDD458 pKa = 4.33GDD460 pKa = 3.55DD461 pKa = 3.63RR462 pKa = 11.84TQVAPHH468 pKa = 6.8AGDD471 pKa = 3.47THH473 pKa = 6.8WYY475 pKa = 9.64GGYY478 pKa = 10.11EE479 pKa = 4.07SQADD483 pKa = 3.45NVLNVDD489 pKa = 4.02AGTSPSTLDD498 pKa = 3.16FWSWNFIEE506 pKa = 4.85QGWDD510 pKa = 3.21YY511 pKa = 11.43GFVEE515 pKa = 4.48ALVGGEE521 pKa = 4.07WVTVPLVDD529 pKa = 5.5DD530 pKa = 4.52EE531 pKa = 5.38GNTVTTDD538 pKa = 4.28DD539 pKa = 5.04DD540 pKa = 3.83PHH542 pKa = 7.9GNNTEE547 pKa = 4.32GNGLTGTSGGEE558 pKa = 4.02YY559 pKa = 10.35FVDD562 pKa = 3.79DD563 pKa = 4.16PVYY566 pKa = 11.09VHH568 pKa = 6.72LTAQLPAGATDD579 pKa = 2.98VRR581 pKa = 11.84FRR583 pKa = 11.84YY584 pKa = 8.58STDD587 pKa = 2.83AAYY590 pKa = 10.92LDD592 pKa = 4.21TGWFVDD598 pKa = 3.97DD599 pKa = 3.96VRR601 pKa = 11.84VDD603 pKa = 3.47GAEE606 pKa = 3.9ASVDD610 pKa = 3.19SDD612 pKa = 3.28AGHH615 pKa = 6.24WTRR618 pKa = 11.84TTGVQEE624 pKa = 4.32NNWTVQTIAACDD636 pKa = 3.85LTPGTTLPGEE646 pKa = 4.47SSDD649 pKa = 3.6GAGNHH654 pKa = 5.53VYY656 pKa = 10.39RR657 pKa = 11.84FEE659 pKa = 6.17GDD661 pKa = 3.1AFEE664 pKa = 4.67QGGLEE669 pKa = 4.23TKK671 pKa = 10.18CANGNQDD678 pKa = 4.15DD679 pKa = 4.25FAVTISNLPTGDD691 pKa = 3.92LTFLDD696 pKa = 3.43ADD698 pKa = 3.79YY699 pKa = 10.31TFRR702 pKa = 11.84VANTGNTKK710 pKa = 10.52
Molecular weight: 76.43 kDa
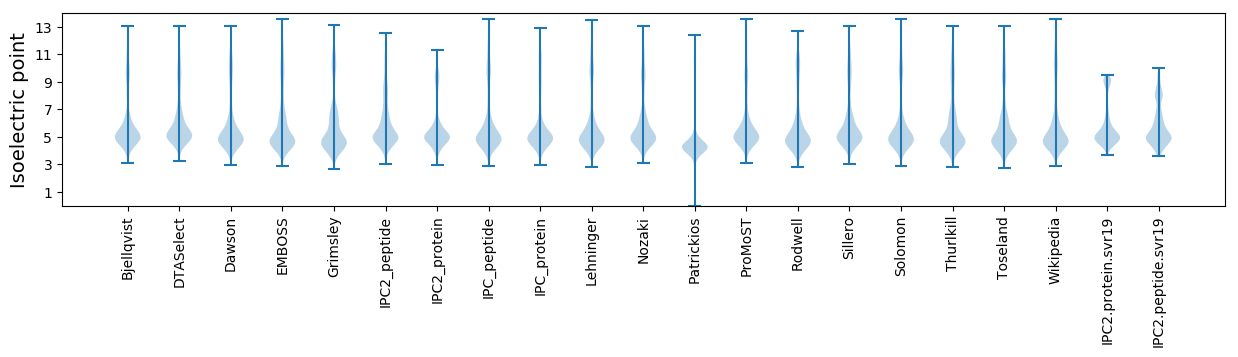
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P8DWF0|A0A2P8DWF0_9ACTN Argininosuccinate lyase OS=Haloactinopolyspora alba OX=648780 GN=CLV30_11334 PE=4 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SIVTSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.7GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SIVTSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.7GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669478 |
25 |
3281 |
323.6 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.108 ± 0.049 | 0.659 ± 0.009 |
7.074 ± 0.033 | 5.898 ± 0.032 |
2.74 ± 0.019 | 9.31 ± 0.034 |
2.27 ± 0.016 | 3.217 ± 0.023 |
1.446 ± 0.02 | 9.622 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.852 ± 0.013 | 1.8 ± 0.02 |
5.741 ± 0.027 | 2.689 ± 0.017 |
8.007 ± 0.039 | 5.284 ± 0.021 |
6.25 ± 0.028 | 9.431 ± 0.038 |
1.558 ± 0.015 | 2.044 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
