
Aureimonas ureilytica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aureimonas
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
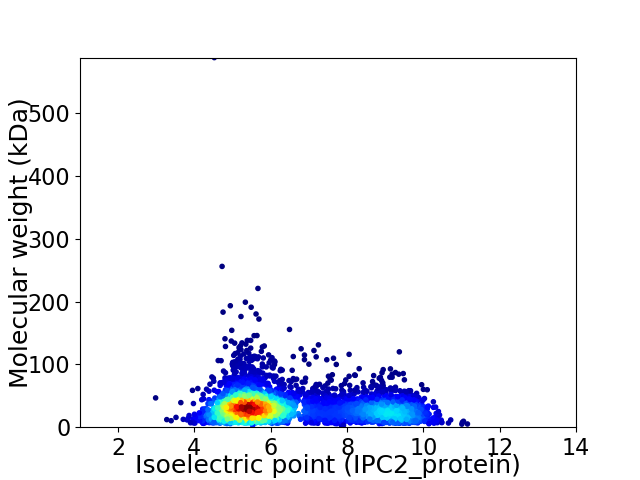
Virtual 2D-PAGE plot for 4329 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
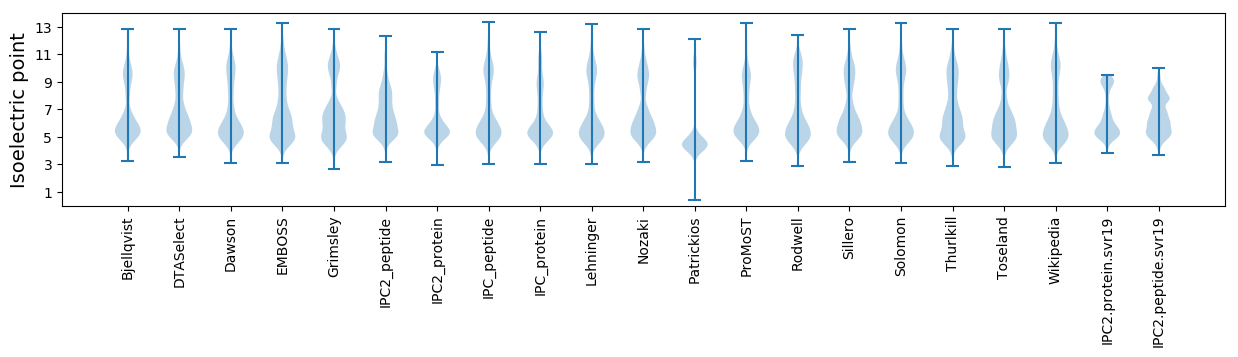
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A175RRL6|A0A175RRL6_9RHIZ ABC transporter permease OS=Aureimonas ureilytica OX=401562 GN=NS365_07690 PE=4 SV=1
MM1 pKa = 7.44SVFKK5 pKa = 10.38STTDD9 pKa = 2.68ATTALKK15 pKa = 9.58FAQMAQAAYY24 pKa = 8.93YY25 pKa = 10.74DD26 pKa = 3.98WMVPPAGWTAVTMGQLGLGNGSNDD50 pKa = 3.93ASTVWHH56 pKa = 6.42GAFGTGAQARR66 pKa = 11.84VFTNGTEE73 pKa = 4.1VTVAIAGTQTDD84 pKa = 4.31NYY86 pKa = 9.73QMGYY90 pKa = 9.87ADD92 pKa = 3.25WMQNVRR98 pKa = 11.84VGQGLTDD105 pKa = 3.45YY106 pKa = 10.5KK107 pKa = 11.28LNFIALVDD115 pKa = 3.75AVKK118 pKa = 9.93TFSEE122 pKa = 4.98APATDD127 pKa = 3.7LAVSFTGHH135 pKa = 6.19SLGAAAVNQIAEE147 pKa = 4.46SNAFGQLPSTTKK159 pKa = 10.48YY160 pKa = 10.73YY161 pKa = 10.91GFEE164 pKa = 3.89SPYY167 pKa = 10.64VFNPPASHH175 pKa = 5.99QAHH178 pKa = 6.34GDD180 pKa = 3.5VMNWGVEE187 pKa = 3.85FDD189 pKa = 4.06LVSGLGLGSGNPEE202 pKa = 3.92YY203 pKa = 10.92GQNIYY208 pKa = 10.64LKK210 pKa = 10.22SDD212 pKa = 3.92FGLTSDD218 pKa = 4.65LFTVHH223 pKa = 7.05SIDD226 pKa = 3.43TQVRR230 pKa = 11.84IIEE233 pKa = 4.68TINKK237 pKa = 9.47SAFFTEE243 pKa = 4.78LNTPATPNEE252 pKa = 4.24SPSTVFITDD261 pKa = 2.98QYY263 pKa = 11.83GFGNHH268 pKa = 6.7DD269 pKa = 3.88FSGSIPMDD277 pKa = 3.51GAYY280 pKa = 9.67IIGSNNWEE288 pKa = 4.46RR289 pKa = 11.84ITGSIHH295 pKa = 7.94DD296 pKa = 4.55DD297 pKa = 4.05RR298 pKa = 11.84IDD300 pKa = 3.69GGGGNDD306 pKa = 4.16IIIGGAGEE314 pKa = 5.01DD315 pKa = 3.4IVAGGGGADD324 pKa = 3.22TFIFSSVPVDD334 pKa = 3.22GLPNVFSPEE343 pKa = 3.72LDD345 pKa = 3.54FVKK348 pKa = 10.38DD349 pKa = 3.96FQVMAYY355 pKa = 9.47EE356 pKa = 4.99GQWNWNPDD364 pKa = 2.91TMEE367 pKa = 4.13WVWQGGFIKK376 pKa = 10.58DD377 pKa = 4.15VIDD380 pKa = 3.59LTAVRR385 pKa = 11.84GIADD389 pKa = 3.71FQGFAAALEE398 pKa = 4.23DD399 pKa = 3.7TAAGLRR405 pKa = 11.84IEE407 pKa = 4.99GSHH410 pKa = 5.98DD411 pKa = 3.52TIYY414 pKa = 10.96LQGVTKK420 pKa = 11.02LEE422 pKa = 4.14MLGSFQDD429 pKa = 3.49LGGGFMFSEE438 pKa = 4.15NVRR441 pKa = 11.84YY442 pKa = 9.95NSSPYY447 pKa = 9.89MSGNPYY453 pKa = 9.37MSSEE457 pKa = 4.15INQAMTGQINGYY469 pKa = 10.12DD470 pKa = 3.99DD471 pKa = 3.75NTLSSEE477 pKa = 4.34LVYY480 pKa = 11.08SADD483 pKa = 3.52PVNNPLGFSIASDD496 pKa = 3.73GTYY499 pKa = 8.15TWSYY503 pKa = 10.06SGPPIPTDD511 pKa = 3.59GSAYY515 pKa = 10.1GYY517 pKa = 10.93AEE519 pKa = 4.21EE520 pKa = 4.24AHH522 pKa = 5.48VTIIDD527 pKa = 3.72KK528 pKa = 11.09YY529 pKa = 9.61GAEE532 pKa = 4.23SDD534 pKa = 3.59VTLYY538 pKa = 9.49GTFYY542 pKa = 10.7SYY544 pKa = 11.6GMAA547 pKa = 4.36
MM1 pKa = 7.44SVFKK5 pKa = 10.38STTDD9 pKa = 2.68ATTALKK15 pKa = 9.58FAQMAQAAYY24 pKa = 8.93YY25 pKa = 10.74DD26 pKa = 3.98WMVPPAGWTAVTMGQLGLGNGSNDD50 pKa = 3.93ASTVWHH56 pKa = 6.42GAFGTGAQARR66 pKa = 11.84VFTNGTEE73 pKa = 4.1VTVAIAGTQTDD84 pKa = 4.31NYY86 pKa = 9.73QMGYY90 pKa = 9.87ADD92 pKa = 3.25WMQNVRR98 pKa = 11.84VGQGLTDD105 pKa = 3.45YY106 pKa = 10.5KK107 pKa = 11.28LNFIALVDD115 pKa = 3.75AVKK118 pKa = 9.93TFSEE122 pKa = 4.98APATDD127 pKa = 3.7LAVSFTGHH135 pKa = 6.19SLGAAAVNQIAEE147 pKa = 4.46SNAFGQLPSTTKK159 pKa = 10.48YY160 pKa = 10.73YY161 pKa = 10.91GFEE164 pKa = 3.89SPYY167 pKa = 10.64VFNPPASHH175 pKa = 5.99QAHH178 pKa = 6.34GDD180 pKa = 3.5VMNWGVEE187 pKa = 3.85FDD189 pKa = 4.06LVSGLGLGSGNPEE202 pKa = 3.92YY203 pKa = 10.92GQNIYY208 pKa = 10.64LKK210 pKa = 10.22SDD212 pKa = 3.92FGLTSDD218 pKa = 4.65LFTVHH223 pKa = 7.05SIDD226 pKa = 3.43TQVRR230 pKa = 11.84IIEE233 pKa = 4.68TINKK237 pKa = 9.47SAFFTEE243 pKa = 4.78LNTPATPNEE252 pKa = 4.24SPSTVFITDD261 pKa = 2.98QYY263 pKa = 11.83GFGNHH268 pKa = 6.7DD269 pKa = 3.88FSGSIPMDD277 pKa = 3.51GAYY280 pKa = 9.67IIGSNNWEE288 pKa = 4.46RR289 pKa = 11.84ITGSIHH295 pKa = 7.94DD296 pKa = 4.55DD297 pKa = 4.05RR298 pKa = 11.84IDD300 pKa = 3.69GGGGNDD306 pKa = 4.16IIIGGAGEE314 pKa = 5.01DD315 pKa = 3.4IVAGGGGADD324 pKa = 3.22TFIFSSVPVDD334 pKa = 3.22GLPNVFSPEE343 pKa = 3.72LDD345 pKa = 3.54FVKK348 pKa = 10.38DD349 pKa = 3.96FQVMAYY355 pKa = 9.47EE356 pKa = 4.99GQWNWNPDD364 pKa = 2.91TMEE367 pKa = 4.13WVWQGGFIKK376 pKa = 10.58DD377 pKa = 4.15VIDD380 pKa = 3.59LTAVRR385 pKa = 11.84GIADD389 pKa = 3.71FQGFAAALEE398 pKa = 4.23DD399 pKa = 3.7TAAGLRR405 pKa = 11.84IEE407 pKa = 4.99GSHH410 pKa = 5.98DD411 pKa = 3.52TIYY414 pKa = 10.96LQGVTKK420 pKa = 11.02LEE422 pKa = 4.14MLGSFQDD429 pKa = 3.49LGGGFMFSEE438 pKa = 4.15NVRR441 pKa = 11.84YY442 pKa = 9.95NSSPYY447 pKa = 9.89MSGNPYY453 pKa = 9.37MSSEE457 pKa = 4.15INQAMTGQINGYY469 pKa = 10.12DD470 pKa = 3.99DD471 pKa = 3.75NTLSSEE477 pKa = 4.34LVYY480 pKa = 11.08SADD483 pKa = 3.52PVNNPLGFSIASDD496 pKa = 3.73GTYY499 pKa = 8.15TWSYY503 pKa = 10.06SGPPIPTDD511 pKa = 3.59GSAYY515 pKa = 10.1GYY517 pKa = 10.93AEE519 pKa = 4.21EE520 pKa = 4.24AHH522 pKa = 5.48VTIIDD527 pKa = 3.72KK528 pKa = 11.09YY529 pKa = 9.61GAEE532 pKa = 4.23SDD534 pKa = 3.59VTLYY538 pKa = 9.49GTFYY542 pKa = 10.7SYY544 pKa = 11.6GMAA547 pKa = 4.36
Molecular weight: 58.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A175RIC6|A0A175RIC6_9RHIZ GDP-mannose 4 6-dehydratase OS=Aureimonas ureilytica OX=401562 GN=gmd PE=3 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84SNHH5 pKa = 5.7LRR7 pKa = 11.84PPRR10 pKa = 11.84GRR12 pKa = 11.84LHH14 pKa = 6.13RR15 pKa = 11.84TLHH18 pKa = 6.02RR19 pKa = 11.84RR20 pKa = 11.84GNPLLILHH28 pKa = 6.61LRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LPQNPPSRR40 pKa = 11.84PLASDD45 pKa = 3.36RR46 pKa = 11.84HH47 pKa = 5.68RR48 pKa = 11.84PSDD51 pKa = 3.31QRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SLTHH59 pKa = 5.35RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.16RR65 pKa = 11.84PPATRR70 pKa = 11.84QPRR73 pKa = 11.84PPRR76 pKa = 11.84PTT78 pKa = 3.13
MM1 pKa = 7.91RR2 pKa = 11.84SNHH5 pKa = 5.7LRR7 pKa = 11.84PPRR10 pKa = 11.84GRR12 pKa = 11.84LHH14 pKa = 6.13RR15 pKa = 11.84TLHH18 pKa = 6.02RR19 pKa = 11.84RR20 pKa = 11.84GNPLLILHH28 pKa = 6.61LRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LPQNPPSRR40 pKa = 11.84PLASDD45 pKa = 3.36RR46 pKa = 11.84HH47 pKa = 5.68RR48 pKa = 11.84PSDD51 pKa = 3.31QRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SLTHH59 pKa = 5.35RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.16RR65 pKa = 11.84PPATRR70 pKa = 11.84QPRR73 pKa = 11.84PPRR76 pKa = 11.84PTT78 pKa = 3.13
Molecular weight: 9.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1349042 |
27 |
5832 |
311.6 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.812 ± 0.042 | 0.763 ± 0.011 |
5.615 ± 0.032 | 5.895 ± 0.037 |
3.816 ± 0.026 | 8.775 ± 0.037 |
1.957 ± 0.018 | 4.907 ± 0.028 |
2.878 ± 0.031 | 10.55 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.019 | 2.35 ± 0.025 |
5.231 ± 0.034 | 2.983 ± 0.025 |
7.611 ± 0.041 | 5.662 ± 0.03 |
5.181 ± 0.022 | 7.367 ± 0.033 |
1.244 ± 0.015 | 2.037 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
