
Pacific flying fox associated cyclovirus-1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Bat associated cyclovirus 14
Average proteome isoelectric point is 8.8
Get precalculated fractions of proteins
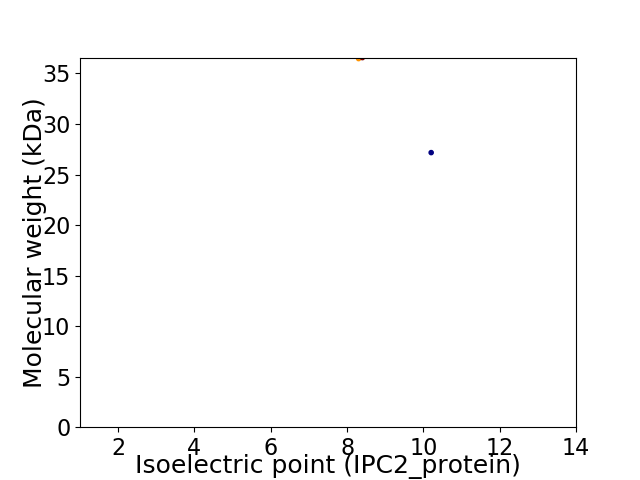
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
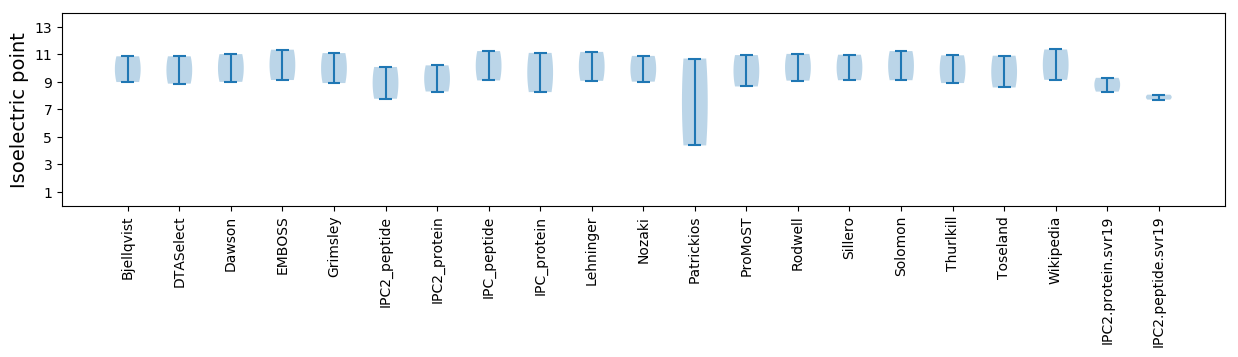
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTJ3|A0A140CTJ3_9CIRC ATP-dependent helicase Rep OS=Pacific flying fox associated cyclovirus-1 OX=1795983 PE=3 SV=1
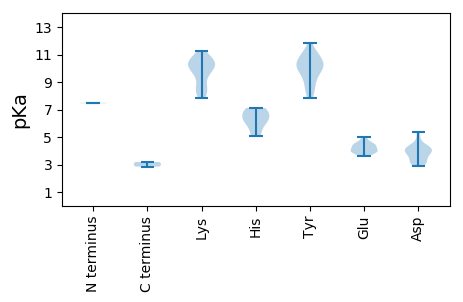
MM1 pKa = 7.45PGEE4 pKa = 4.51PKK6 pKa = 9.57SHH8 pKa = 6.0RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.77VFTLNNYY18 pKa = 10.04SDD20 pKa = 5.35DD21 pKa = 3.85DD22 pKa = 4.03VARR25 pKa = 11.84LQQLEE30 pKa = 3.92ARR32 pKa = 11.84YY33 pKa = 8.3MVFGKK38 pKa = 10.25EE39 pKa = 3.92VAPTSGTKK47 pKa = 10.01HH48 pKa = 5.3LQGFINFGRR57 pKa = 11.84AVRR60 pKa = 11.84FNSARR65 pKa = 11.84KK66 pKa = 9.12LVGGDD71 pKa = 3.08GVTEE75 pKa = 4.2VTDD78 pKa = 4.23RR79 pKa = 11.84AASGGPNGDD88 pKa = 3.73CFDD91 pKa = 4.35YY92 pKa = 10.84LPHH95 pKa = 7.02GNDD98 pKa = 3.08PATRR102 pKa = 11.84GFVFLGKK109 pKa = 8.46PTSQGRR115 pKa = 11.84RR116 pKa = 11.84TDD118 pKa = 3.57LQRR121 pKa = 11.84VADD124 pKa = 4.17LAKK127 pKa = 10.58QSGTTAHH134 pKa = 6.74TIARR138 pKa = 11.84EE139 pKa = 3.94FPVEE143 pKa = 4.29FIKK146 pKa = 10.89YY147 pKa = 9.94GRR149 pKa = 11.84GITNLLRR156 pKa = 11.84TINPIPSRR164 pKa = 11.84SKK166 pKa = 8.04PTIVVVLVGAPGVGKK181 pKa = 10.52SRR183 pKa = 11.84FANEE187 pKa = 3.94VGSGLGDD194 pKa = 3.5TYY196 pKa = 11.27YY197 pKa = 10.95KK198 pKa = 10.51PRR200 pKa = 11.84GEE202 pKa = 3.94WWDD205 pKa = 4.56GYY207 pKa = 9.1CQQRR211 pKa = 11.84SVIIDD216 pKa = 3.92DD217 pKa = 4.33FYY219 pKa = 11.87GWIKK223 pKa = 10.75YY224 pKa = 10.28DD225 pKa = 4.1EE226 pKa = 4.44LLKK229 pKa = 10.69ICDD232 pKa = 4.0RR233 pKa = 11.84YY234 pKa = 8.26PHH236 pKa = 6.41KK237 pKa = 11.26VPVKK241 pKa = 9.99GGYY244 pKa = 8.95EE245 pKa = 4.19EE246 pKa = 4.38FTSEE250 pKa = 4.47YY251 pKa = 11.25VFITSNSNVDD261 pKa = 2.88QWYY264 pKa = 9.92KK265 pKa = 10.4FDD267 pKa = 4.24NYY269 pKa = 10.85SPAAIHH275 pKa = 6.52RR276 pKa = 11.84RR277 pKa = 11.84LSVYY281 pKa = 10.22VGLMCDD287 pKa = 3.1GSTAHH292 pKa = 6.16FPEE295 pKa = 4.65WSPKK299 pKa = 10.09FSWFDD304 pKa = 3.72SAWTTMQIKK313 pKa = 7.8WTVHH317 pKa = 6.03DD318 pKa = 4.31MVNKK322 pKa = 9.82VV323 pKa = 2.85
MM1 pKa = 7.45PGEE4 pKa = 4.51PKK6 pKa = 9.57SHH8 pKa = 6.0RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.77VFTLNNYY18 pKa = 10.04SDD20 pKa = 5.35DD21 pKa = 3.85DD22 pKa = 4.03VARR25 pKa = 11.84LQQLEE30 pKa = 3.92ARR32 pKa = 11.84YY33 pKa = 8.3MVFGKK38 pKa = 10.25EE39 pKa = 3.92VAPTSGTKK47 pKa = 10.01HH48 pKa = 5.3LQGFINFGRR57 pKa = 11.84AVRR60 pKa = 11.84FNSARR65 pKa = 11.84KK66 pKa = 9.12LVGGDD71 pKa = 3.08GVTEE75 pKa = 4.2VTDD78 pKa = 4.23RR79 pKa = 11.84AASGGPNGDD88 pKa = 3.73CFDD91 pKa = 4.35YY92 pKa = 10.84LPHH95 pKa = 7.02GNDD98 pKa = 3.08PATRR102 pKa = 11.84GFVFLGKK109 pKa = 8.46PTSQGRR115 pKa = 11.84RR116 pKa = 11.84TDD118 pKa = 3.57LQRR121 pKa = 11.84VADD124 pKa = 4.17LAKK127 pKa = 10.58QSGTTAHH134 pKa = 6.74TIARR138 pKa = 11.84EE139 pKa = 3.94FPVEE143 pKa = 4.29FIKK146 pKa = 10.89YY147 pKa = 9.94GRR149 pKa = 11.84GITNLLRR156 pKa = 11.84TINPIPSRR164 pKa = 11.84SKK166 pKa = 8.04PTIVVVLVGAPGVGKK181 pKa = 10.52SRR183 pKa = 11.84FANEE187 pKa = 3.94VGSGLGDD194 pKa = 3.5TYY196 pKa = 11.27YY197 pKa = 10.95KK198 pKa = 10.51PRR200 pKa = 11.84GEE202 pKa = 3.94WWDD205 pKa = 4.56GYY207 pKa = 9.1CQQRR211 pKa = 11.84SVIIDD216 pKa = 3.92DD217 pKa = 4.33FYY219 pKa = 11.87GWIKK223 pKa = 10.75YY224 pKa = 10.28DD225 pKa = 4.1EE226 pKa = 4.44LLKK229 pKa = 10.69ICDD232 pKa = 4.0RR233 pKa = 11.84YY234 pKa = 8.26PHH236 pKa = 6.41KK237 pKa = 11.26VPVKK241 pKa = 9.99GGYY244 pKa = 8.95EE245 pKa = 4.19EE246 pKa = 4.38FTSEE250 pKa = 4.47YY251 pKa = 11.25VFITSNSNVDD261 pKa = 2.88QWYY264 pKa = 9.92KK265 pKa = 10.4FDD267 pKa = 4.24NYY269 pKa = 10.85SPAAIHH275 pKa = 6.52RR276 pKa = 11.84RR277 pKa = 11.84LSVYY281 pKa = 10.22VGLMCDD287 pKa = 3.1GSTAHH292 pKa = 6.16FPEE295 pKa = 4.65WSPKK299 pKa = 10.09FSWFDD304 pKa = 3.72SAWTTMQIKK313 pKa = 7.8WTVHH317 pKa = 6.03DD318 pKa = 4.31MVNKK322 pKa = 9.82VV323 pKa = 2.85
Molecular weight: 36.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTJ3|A0A140CTJ3_9CIRC ATP-dependent helicase Rep OS=Pacific flying fox associated cyclovirus-1 OX=1795983 PE=3 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84MRR5 pKa = 11.84LRR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 8.76RR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84VVAPRR18 pKa = 11.84RR19 pKa = 11.84GIRR22 pKa = 11.84KK23 pKa = 8.38RR24 pKa = 11.84RR25 pKa = 11.84SFRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84NRR33 pKa = 11.84KK34 pKa = 8.41YY35 pKa = 10.11DD36 pKa = 3.24VHH38 pKa = 6.88NFRR41 pKa = 11.84LKK43 pKa = 9.93SGSYY47 pKa = 10.3VVDD50 pKa = 3.85LTPGVNGGARR60 pKa = 11.84VVGEE64 pKa = 3.88AVVLNQLPINFNGLTLLYY82 pKa = 10.32SHH84 pKa = 7.11IRR86 pKa = 11.84ITKK89 pKa = 8.4AVYY92 pKa = 10.12RR93 pKa = 11.84FIPMYY98 pKa = 7.83TQRR101 pKa = 11.84TWPDD105 pKa = 3.2PATNDD110 pKa = 3.06TDD112 pKa = 3.91LPEE115 pKa = 4.98ILSTQWYY122 pKa = 9.47GPALDD127 pKa = 4.14VDD129 pKa = 4.98SNPSQSYY136 pKa = 9.95NVLRR140 pKa = 11.84SDD142 pKa = 4.12FLTRR146 pKa = 11.84NHH148 pKa = 5.1QWNRR152 pKa = 11.84PFKK155 pKa = 9.97RR156 pKa = 11.84VLYY159 pKa = 9.35PKK161 pKa = 10.39VNYY164 pKa = 9.51FVTASTYY171 pKa = 10.69VDD173 pKa = 3.19NAAPNSNTACALKK186 pKa = 10.43KK187 pKa = 9.69MPWVRR192 pKa = 11.84TSGSNAYY199 pKa = 10.33DD200 pKa = 3.63NLQCGVMYY208 pKa = 10.44VGAIGGPAGAAVKK221 pKa = 10.48YY222 pKa = 9.75RR223 pKa = 11.84VEE225 pKa = 3.6RR226 pKa = 11.84TYY228 pKa = 11.0YY229 pKa = 8.42VQARR233 pKa = 11.84KK234 pKa = 8.87VV235 pKa = 3.19
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84MRR5 pKa = 11.84LRR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 8.76RR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84VVAPRR18 pKa = 11.84RR19 pKa = 11.84GIRR22 pKa = 11.84KK23 pKa = 8.38RR24 pKa = 11.84RR25 pKa = 11.84SFRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84NRR33 pKa = 11.84KK34 pKa = 8.41YY35 pKa = 10.11DD36 pKa = 3.24VHH38 pKa = 6.88NFRR41 pKa = 11.84LKK43 pKa = 9.93SGSYY47 pKa = 10.3VVDD50 pKa = 3.85LTPGVNGGARR60 pKa = 11.84VVGEE64 pKa = 3.88AVVLNQLPINFNGLTLLYY82 pKa = 10.32SHH84 pKa = 7.11IRR86 pKa = 11.84ITKK89 pKa = 8.4AVYY92 pKa = 10.12RR93 pKa = 11.84FIPMYY98 pKa = 7.83TQRR101 pKa = 11.84TWPDD105 pKa = 3.2PATNDD110 pKa = 3.06TDD112 pKa = 3.91LPEE115 pKa = 4.98ILSTQWYY122 pKa = 9.47GPALDD127 pKa = 4.14VDD129 pKa = 4.98SNPSQSYY136 pKa = 9.95NVLRR140 pKa = 11.84SDD142 pKa = 4.12FLTRR146 pKa = 11.84NHH148 pKa = 5.1QWNRR152 pKa = 11.84PFKK155 pKa = 9.97RR156 pKa = 11.84VLYY159 pKa = 9.35PKK161 pKa = 10.39VNYY164 pKa = 9.51FVTASTYY171 pKa = 10.69VDD173 pKa = 3.19NAAPNSNTACALKK186 pKa = 10.43KK187 pKa = 9.69MPWVRR192 pKa = 11.84TSGSNAYY199 pKa = 10.33DD200 pKa = 3.63NLQCGVMYY208 pKa = 10.44VGAIGGPAGAAVKK221 pKa = 10.48YY222 pKa = 9.75RR223 pKa = 11.84VEE225 pKa = 3.6RR226 pKa = 11.84TYY228 pKa = 11.0YY229 pKa = 8.42VQARR233 pKa = 11.84KK234 pKa = 8.87VV235 pKa = 3.19
Molecular weight: 27.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
558 |
235 |
323 |
279.0 |
31.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.272 ± 0.569 | 1.075 ± 0.133 |
5.556 ± 0.769 | 2.867 ± 0.941 |
4.48 ± 0.888 | 8.065 ± 1.247 |
1.971 ± 0.411 | 3.763 ± 0.464 |
5.018 ± 0.451 | 5.914 ± 0.529 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.792 ± 0.199 | 5.376 ± 1.099 |
5.914 ± 0.277 | 3.047 ± 0.04 |
9.498 ± 2.185 | 6.093 ± 0.332 |
6.272 ± 0.186 | 9.14 ± 0.635 |
2.151 ± 0.265 | 5.735 ± 0.635 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
