
Colletotrichum gloeosporioides (strain Cg-14) (Anthracnose fungus) (Glomerella cingulata)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Glomerellales; Glomerellaceae; Colletotrichum; Colletotrichum gloeosporioides species complex; Colletotrichum gloeosporioides
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 16388 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
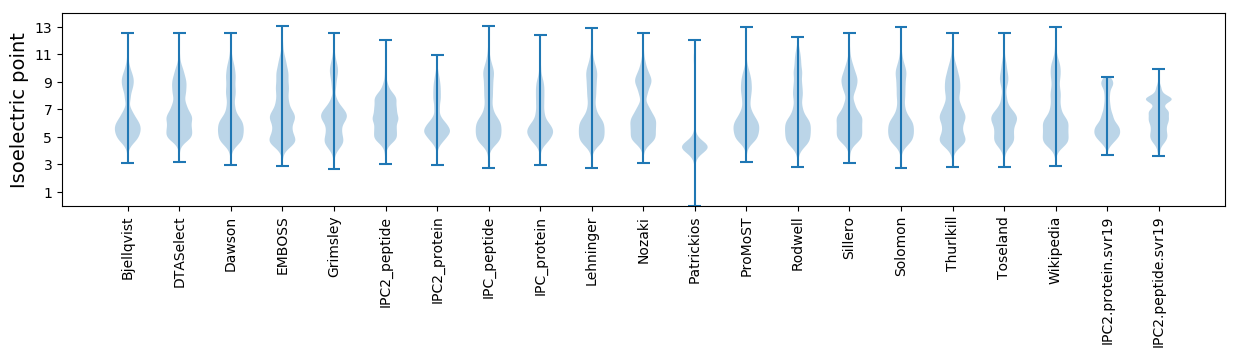
* You can choose from 21 different methods for calculating isoelectric point
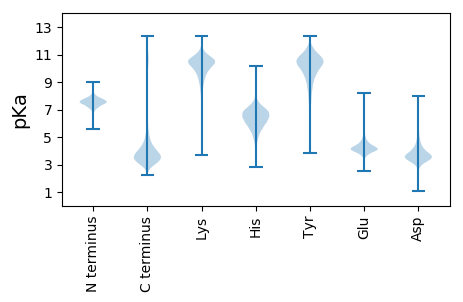
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T0KNM6|T0KNM6_COLGC Protein kinase OS=Colletotrichum gloeosporioides (strain Cg-14) OX=1237896 GN=CGLO_06083 PE=4 SV=1
MM1 pKa = 7.57ANVPVTIDD9 pKa = 3.13MDD11 pKa = 4.21LGSNCLQDD19 pKa = 3.73PSDD22 pKa = 4.01YY23 pKa = 10.73STDD26 pKa = 3.85DD27 pKa = 3.24LTSYY31 pKa = 9.43YY32 pKa = 10.72AAYY35 pKa = 7.98RR36 pKa = 11.84TTSLTDD42 pKa = 3.15SFGSDD47 pKa = 2.93FSTYY51 pKa = 9.16QDD53 pKa = 3.4YY54 pKa = 11.56LDD56 pKa = 4.07YY57 pKa = 10.64YY58 pKa = 11.14KK59 pKa = 10.65KK60 pKa = 9.39VTQIGDD66 pKa = 3.18IGSALANMDD75 pKa = 3.51GQHH78 pKa = 6.28TLDD81 pKa = 4.46AYY83 pKa = 11.38NPDD86 pKa = 3.55EE87 pKa = 4.38SLTPSFLTFQTEE99 pKa = 3.63IDD101 pKa = 3.46GGLFDD106 pKa = 4.7GCIGSHH112 pKa = 5.37SSKK115 pKa = 9.73TFSLDD120 pKa = 2.92VDD122 pKa = 3.74GTSYY126 pKa = 11.03QITLEE131 pKa = 4.66DD132 pKa = 4.6PPTSQWTITQDD143 pKa = 3.81SPGDD147 pKa = 3.56PNRR150 pKa = 11.84IVIGSGGRR158 pKa = 11.84ANPLLTVLYY167 pKa = 9.96AVLGVEE173 pKa = 4.4ILALAGAVLGSALGGLAPAAVEE195 pKa = 4.07GTIAEE200 pKa = 4.37MGTSEE205 pKa = 4.06MTQIIEE211 pKa = 4.8DD212 pKa = 4.26KK213 pKa = 10.48LASDD217 pKa = 4.22PWVMQWMEE225 pKa = 3.72KK226 pKa = 10.05TIFGEE231 pKa = 4.3AEE233 pKa = 3.44AAAKK237 pKa = 10.05KK238 pKa = 9.84RR239 pKa = 11.84ASPVARR245 pKa = 11.84RR246 pKa = 11.84GVGAGGILQEE256 pKa = 3.83WTAAYY261 pKa = 7.59DD262 pKa = 3.12TWNYY266 pKa = 10.52DD267 pKa = 3.64DD268 pKa = 6.03RR269 pKa = 11.84DD270 pKa = 3.87ACVMSKK276 pKa = 10.91DD277 pKa = 3.87VIGEE281 pKa = 4.17LDD283 pKa = 3.95CLVVGTSIVIQADD296 pKa = 4.2GQPMAMPLPPLGSAWW311 pKa = 3.32
MM1 pKa = 7.57ANVPVTIDD9 pKa = 3.13MDD11 pKa = 4.21LGSNCLQDD19 pKa = 3.73PSDD22 pKa = 4.01YY23 pKa = 10.73STDD26 pKa = 3.85DD27 pKa = 3.24LTSYY31 pKa = 9.43YY32 pKa = 10.72AAYY35 pKa = 7.98RR36 pKa = 11.84TTSLTDD42 pKa = 3.15SFGSDD47 pKa = 2.93FSTYY51 pKa = 9.16QDD53 pKa = 3.4YY54 pKa = 11.56LDD56 pKa = 4.07YY57 pKa = 10.64YY58 pKa = 11.14KK59 pKa = 10.65KK60 pKa = 9.39VTQIGDD66 pKa = 3.18IGSALANMDD75 pKa = 3.51GQHH78 pKa = 6.28TLDD81 pKa = 4.46AYY83 pKa = 11.38NPDD86 pKa = 3.55EE87 pKa = 4.38SLTPSFLTFQTEE99 pKa = 3.63IDD101 pKa = 3.46GGLFDD106 pKa = 4.7GCIGSHH112 pKa = 5.37SSKK115 pKa = 9.73TFSLDD120 pKa = 2.92VDD122 pKa = 3.74GTSYY126 pKa = 11.03QITLEE131 pKa = 4.66DD132 pKa = 4.6PPTSQWTITQDD143 pKa = 3.81SPGDD147 pKa = 3.56PNRR150 pKa = 11.84IVIGSGGRR158 pKa = 11.84ANPLLTVLYY167 pKa = 9.96AVLGVEE173 pKa = 4.4ILALAGAVLGSALGGLAPAAVEE195 pKa = 4.07GTIAEE200 pKa = 4.37MGTSEE205 pKa = 4.06MTQIIEE211 pKa = 4.8DD212 pKa = 4.26KK213 pKa = 10.48LASDD217 pKa = 4.22PWVMQWMEE225 pKa = 3.72KK226 pKa = 10.05TIFGEE231 pKa = 4.3AEE233 pKa = 3.44AAAKK237 pKa = 10.05KK238 pKa = 9.84RR239 pKa = 11.84ASPVARR245 pKa = 11.84RR246 pKa = 11.84GVGAGGILQEE256 pKa = 3.83WTAAYY261 pKa = 7.59DD262 pKa = 3.12TWNYY266 pKa = 10.52DD267 pKa = 3.64DD268 pKa = 6.03RR269 pKa = 11.84DD270 pKa = 3.87ACVMSKK276 pKa = 10.91DD277 pKa = 3.87VIGEE281 pKa = 4.17LDD283 pKa = 3.95CLVVGTSIVIQADD296 pKa = 4.2GQPMAMPLPPLGSAWW311 pKa = 3.32
Molecular weight: 33.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T0LYN2|T0LYN2_COLGC Uncharacterized protein OS=Colletotrichum gloeosporioides (strain Cg-14) OX=1237896 GN=CGLO_03306 PE=4 SV=1
MM1 pKa = 7.85PLRR4 pKa = 11.84STRR7 pKa = 11.84HH8 pKa = 4.05TTTTAPRR15 pKa = 11.84RR16 pKa = 11.84GGLFTRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84APARR28 pKa = 11.84THH30 pKa = 5.82HH31 pKa = 6.33HH32 pKa = 6.15HH33 pKa = 6.18TTTTTTTRR41 pKa = 11.84KK42 pKa = 9.94RR43 pKa = 11.84GGLFSRR49 pKa = 11.84RR50 pKa = 11.84HH51 pKa = 5.06GAVTTAPVHH60 pKa = 4.81HH61 pKa = 6.22QRR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.81PSMGDD70 pKa = 3.3KK71 pKa = 10.53ISGAMLKK78 pKa = 10.72LKK80 pKa = 9.36GTLTRR85 pKa = 11.84RR86 pKa = 11.84PGQKK90 pKa = 9.9AAGTRR95 pKa = 11.84RR96 pKa = 11.84MHH98 pKa = 5.68GTDD101 pKa = 2.6GRR103 pKa = 11.84GSHH106 pKa = 5.87RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 10.21RR110 pKa = 11.84YY111 pKa = 9.72
MM1 pKa = 7.85PLRR4 pKa = 11.84STRR7 pKa = 11.84HH8 pKa = 4.05TTTTAPRR15 pKa = 11.84RR16 pKa = 11.84GGLFTRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84APARR28 pKa = 11.84THH30 pKa = 5.82HH31 pKa = 6.33HH32 pKa = 6.15HH33 pKa = 6.18TTTTTTTRR41 pKa = 11.84KK42 pKa = 9.94RR43 pKa = 11.84GGLFSRR49 pKa = 11.84RR50 pKa = 11.84HH51 pKa = 5.06GAVTTAPVHH60 pKa = 4.81HH61 pKa = 6.22QRR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.81PSMGDD70 pKa = 3.3KK71 pKa = 10.53ISGAMLKK78 pKa = 10.72LKK80 pKa = 9.36GTLTRR85 pKa = 11.84RR86 pKa = 11.84PGQKK90 pKa = 9.9AAGTRR95 pKa = 11.84RR96 pKa = 11.84MHH98 pKa = 5.68GTDD101 pKa = 2.6GRR103 pKa = 11.84GSHH106 pKa = 5.87RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 10.21RR110 pKa = 11.84YY111 pKa = 9.72
Molecular weight: 12.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6823665 |
8 |
6253 |
416.4 |
46.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.043 ± 0.017 | 1.253 ± 0.008 |
5.82 ± 0.014 | 6.055 ± 0.02 |
3.913 ± 0.012 | 7.144 ± 0.02 |
2.288 ± 0.009 | 4.899 ± 0.014 |
4.847 ± 0.02 | 8.725 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.304 ± 0.007 | 3.728 ± 0.011 |
5.903 ± 0.022 | 3.797 ± 0.014 |
5.775 ± 0.017 | 7.731 ± 0.021 |
5.963 ± 0.018 | 6.371 ± 0.015 |
1.627 ± 0.008 | 2.815 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
