
Fiddler Crab associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.23
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
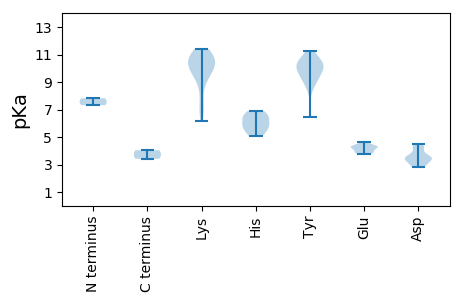
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0K1RLN8|A0A0K1RLN8_9CIRC Putative capsid protein OS=Fiddler Crab associated circular virus OX=1692249 PE=4 SV=1
MM1 pKa = 7.31GVKK4 pKa = 10.47LKK6 pKa = 10.62QSQDD10 pKa = 3.01NSEE13 pKa = 4.46GGNNHH18 pKa = 6.47PPSFRR23 pKa = 11.84SRR25 pKa = 11.84KK26 pKa = 9.64LVFTLNNYY34 pKa = 8.41QKK36 pKa = 11.16EE37 pKa = 4.38DD38 pKa = 3.19INLIISYY45 pKa = 10.13FEE47 pKa = 4.02SSAQKK52 pKa = 9.17WVFGEE57 pKa = 4.19EE58 pKa = 4.22VGEE61 pKa = 4.28SGTPHH66 pKa = 5.89LQGYY70 pKa = 9.1CEE72 pKa = 4.38FKK74 pKa = 10.74NQKK77 pKa = 7.26EE78 pKa = 4.35WKK80 pKa = 9.92QITDD84 pKa = 2.81KK85 pKa = 11.17CSPLKK90 pKa = 10.52RR91 pKa = 11.84AWSTSARR98 pKa = 11.84GSLDD102 pKa = 4.47DD103 pKa = 3.79NVKK106 pKa = 9.22YY107 pKa = 6.46TTKK110 pKa = 10.54DD111 pKa = 2.87GKK113 pKa = 10.33IYY115 pKa = 10.67YY116 pKa = 10.36GGFKK120 pKa = 9.68IEE122 pKa = 3.78KK123 pKa = 8.96CNYY126 pKa = 8.83KK127 pKa = 10.19VEE129 pKa = 4.07IDD131 pKa = 3.51LYY133 pKa = 10.14DD134 pKa = 3.39WQKK137 pKa = 11.38EE138 pKa = 3.75IMEE141 pKa = 4.66ILEE144 pKa = 3.99QTPDD148 pKa = 3.3DD149 pKa = 4.13RR150 pKa = 11.84TIHH153 pKa = 5.7WVWEE157 pKa = 4.09PKK159 pKa = 10.7GGIGKK164 pKa = 7.01TTFQKK169 pKa = 11.15YY170 pKa = 9.24IFTHH174 pKa = 5.08MGGVCVLSGKK184 pKa = 10.37GSDD187 pKa = 3.33MKK189 pKa = 10.8HH190 pKa = 5.3GVVSYY195 pKa = 10.96INDD198 pKa = 3.55HH199 pKa = 6.6GEE201 pKa = 3.92TPDD204 pKa = 3.25IVLINIPRR212 pKa = 11.84SSAEE216 pKa = 3.8FVSWSGIEE224 pKa = 4.16EE225 pKa = 3.94IKK227 pKa = 11.33DD228 pKa = 3.06MFFYY232 pKa = 10.48SGKK235 pKa = 9.79YY236 pKa = 9.55EE237 pKa = 4.23GGQVCGMSPHH247 pKa = 6.07VLVFANTEE255 pKa = 3.81PPEE258 pKa = 4.34GVFSKK263 pKa = 10.99DD264 pKa = 2.88RR265 pKa = 11.84VKK267 pKa = 10.26TIEE270 pKa = 4.26LDD272 pKa = 3.54SS273 pKa = 4.04
MM1 pKa = 7.31GVKK4 pKa = 10.47LKK6 pKa = 10.62QSQDD10 pKa = 3.01NSEE13 pKa = 4.46GGNNHH18 pKa = 6.47PPSFRR23 pKa = 11.84SRR25 pKa = 11.84KK26 pKa = 9.64LVFTLNNYY34 pKa = 8.41QKK36 pKa = 11.16EE37 pKa = 4.38DD38 pKa = 3.19INLIISYY45 pKa = 10.13FEE47 pKa = 4.02SSAQKK52 pKa = 9.17WVFGEE57 pKa = 4.19EE58 pKa = 4.22VGEE61 pKa = 4.28SGTPHH66 pKa = 5.89LQGYY70 pKa = 9.1CEE72 pKa = 4.38FKK74 pKa = 10.74NQKK77 pKa = 7.26EE78 pKa = 4.35WKK80 pKa = 9.92QITDD84 pKa = 2.81KK85 pKa = 11.17CSPLKK90 pKa = 10.52RR91 pKa = 11.84AWSTSARR98 pKa = 11.84GSLDD102 pKa = 4.47DD103 pKa = 3.79NVKK106 pKa = 9.22YY107 pKa = 6.46TTKK110 pKa = 10.54DD111 pKa = 2.87GKK113 pKa = 10.33IYY115 pKa = 10.67YY116 pKa = 10.36GGFKK120 pKa = 9.68IEE122 pKa = 3.78KK123 pKa = 8.96CNYY126 pKa = 8.83KK127 pKa = 10.19VEE129 pKa = 4.07IDD131 pKa = 3.51LYY133 pKa = 10.14DD134 pKa = 3.39WQKK137 pKa = 11.38EE138 pKa = 3.75IMEE141 pKa = 4.66ILEE144 pKa = 3.99QTPDD148 pKa = 3.3DD149 pKa = 4.13RR150 pKa = 11.84TIHH153 pKa = 5.7WVWEE157 pKa = 4.09PKK159 pKa = 10.7GGIGKK164 pKa = 7.01TTFQKK169 pKa = 11.15YY170 pKa = 9.24IFTHH174 pKa = 5.08MGGVCVLSGKK184 pKa = 10.37GSDD187 pKa = 3.33MKK189 pKa = 10.8HH190 pKa = 5.3GVVSYY195 pKa = 10.96INDD198 pKa = 3.55HH199 pKa = 6.6GEE201 pKa = 3.92TPDD204 pKa = 3.25IVLINIPRR212 pKa = 11.84SSAEE216 pKa = 3.8FVSWSGIEE224 pKa = 4.16EE225 pKa = 3.94IKK227 pKa = 11.33DD228 pKa = 3.06MFFYY232 pKa = 10.48SGKK235 pKa = 9.79YY236 pKa = 9.55EE237 pKa = 4.23GGQVCGMSPHH247 pKa = 6.07VLVFANTEE255 pKa = 3.81PPEE258 pKa = 4.34GVFSKK263 pKa = 10.99DD264 pKa = 2.88RR265 pKa = 11.84VKK267 pKa = 10.26TIEE270 pKa = 4.26LDD272 pKa = 3.54SS273 pKa = 4.04
Molecular weight: 31.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLN8|A0A0K1RLN8_9CIRC Putative capsid protein OS=Fiddler Crab associated circular virus OX=1692249 PE=4 SV=1
MM1 pKa = 7.82PYY3 pKa = 10.6ARR5 pKa = 11.84KK6 pKa = 6.87TQKK9 pKa = 9.82SRR11 pKa = 11.84KK12 pKa = 6.16RR13 pKa = 11.84TYY15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 8.02TYY20 pKa = 9.82RR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.47NLVLSKK29 pKa = 11.11APIPNTFSTKK39 pKa = 9.66LRR41 pKa = 11.84YY42 pKa = 9.4SEE44 pKa = 4.38SVSVNPGASGVAGVYY59 pKa = 10.35VMNASSCYY67 pKa = 10.32DD68 pKa = 3.57PNTTGTGHH76 pKa = 5.7QPRR79 pKa = 11.84GFDD82 pKa = 2.85QWMTMYY88 pKa = 11.26DD89 pKa = 3.49HH90 pKa = 6.49FTVIGAKK97 pKa = 8.42ITAQFSITGQADD109 pKa = 3.4DD110 pKa = 4.24RR111 pKa = 11.84ALICGIALKK120 pKa = 10.35DD121 pKa = 3.74ANGVYY126 pKa = 10.35ADD128 pKa = 4.26KK129 pKa = 11.02NDD131 pKa = 3.59YY132 pKa = 10.07MEE134 pKa = 4.28GRR136 pKa = 11.84NVVSKK141 pKa = 10.43LVSGGATGDD150 pKa = 3.38ARR152 pKa = 11.84TVTLSKK158 pKa = 9.79TYY160 pKa = 9.85SAKK163 pKa = 10.5KK164 pKa = 9.71FLGISKK170 pKa = 9.52PLASSLIRR178 pKa = 11.84GTSSANPSEE187 pKa = 3.85NAYY190 pKa = 10.14FHH192 pKa = 6.89LFAAPTGNADD202 pKa = 4.34LDD204 pKa = 4.32NIPIQVLIDD213 pKa = 3.56YY214 pKa = 9.22LVVFTEE220 pKa = 4.53PQQPSQSS227 pKa = 3.42
MM1 pKa = 7.82PYY3 pKa = 10.6ARR5 pKa = 11.84KK6 pKa = 6.87TQKK9 pKa = 9.82SRR11 pKa = 11.84KK12 pKa = 6.16RR13 pKa = 11.84TYY15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 8.02TYY20 pKa = 9.82RR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.47NLVLSKK29 pKa = 11.11APIPNTFSTKK39 pKa = 9.66LRR41 pKa = 11.84YY42 pKa = 9.4SEE44 pKa = 4.38SVSVNPGASGVAGVYY59 pKa = 10.35VMNASSCYY67 pKa = 10.32DD68 pKa = 3.57PNTTGTGHH76 pKa = 5.7QPRR79 pKa = 11.84GFDD82 pKa = 2.85QWMTMYY88 pKa = 11.26DD89 pKa = 3.49HH90 pKa = 6.49FTVIGAKK97 pKa = 8.42ITAQFSITGQADD109 pKa = 3.4DD110 pKa = 4.24RR111 pKa = 11.84ALICGIALKK120 pKa = 10.35DD121 pKa = 3.74ANGVYY126 pKa = 10.35ADD128 pKa = 4.26KK129 pKa = 11.02NDD131 pKa = 3.59YY132 pKa = 10.07MEE134 pKa = 4.28GRR136 pKa = 11.84NVVSKK141 pKa = 10.43LVSGGATGDD150 pKa = 3.38ARR152 pKa = 11.84TVTLSKK158 pKa = 9.79TYY160 pKa = 9.85SAKK163 pKa = 10.5KK164 pKa = 9.71FLGISKK170 pKa = 9.52PLASSLIRR178 pKa = 11.84GTSSANPSEE187 pKa = 3.85NAYY190 pKa = 10.14FHH192 pKa = 6.89LFAAPTGNADD202 pKa = 4.34LDD204 pKa = 4.32NIPIQVLIDD213 pKa = 3.56YY214 pKa = 9.22LVVFTEE220 pKa = 4.53PQQPSQSS227 pKa = 3.42
Molecular weight: 24.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
500 |
227 |
273 |
250.0 |
27.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.2 ± 2.598 | 1.4 ± 0.333 |
5.6 ± 0.201 | 5.2 ± 2.205 |
4.2 ± 0.433 | 8.6 ± 0.712 |
2.0 ± 0.435 | 6.0 ± 0.74 |
8.2 ± 1.021 | 5.4 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.002 | 4.8 ± 0.312 |
4.6 ± 0.44 | 4.0 ± 0.023 |
4.0 ± 1.107 | 9.0 ± 0.444 |
6.6 ± 1.135 | 6.6 ± 0.005 |
1.6 ± 0.744 | 4.8 ± 0.312 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
