
Gastropod associated circular ssDNA virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
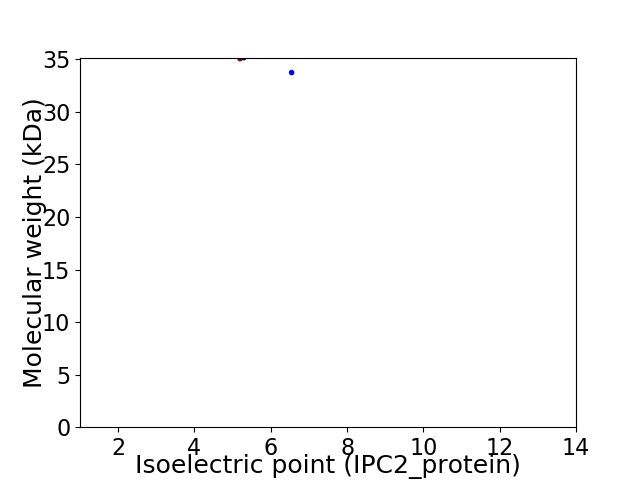
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1RKT4|M1RKT4_9VIRU Putative capsid protein OS=Gastropod associated circular ssDNA virus OX=1296573 PE=4 SV=1
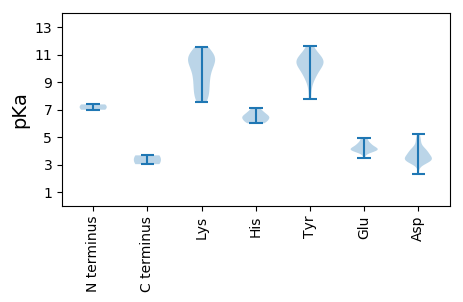
MM1 pKa = 7.37NMAKK5 pKa = 8.92RR6 pKa = 11.84TNKK9 pKa = 8.16THH11 pKa = 6.14AQRR14 pKa = 11.84AFQIDD19 pKa = 4.16TIGGSGQKK27 pKa = 9.76QLIQLDD33 pKa = 3.71ACLSKK38 pKa = 10.91LNHH41 pKa = 6.1RR42 pKa = 11.84LYY44 pKa = 10.74RR45 pKa = 11.84QGRR48 pKa = 11.84MYY50 pKa = 10.25RR51 pKa = 11.84AKK53 pKa = 10.76VNLPASQMPVVNSEE67 pKa = 4.51PFPQFPQTKK76 pKa = 9.71GIEE79 pKa = 3.98VWALMDD85 pKa = 3.43TWYY88 pKa = 10.73VHH90 pKa = 6.82AMWKK94 pKa = 7.54QAKK97 pKa = 9.06RR98 pKa = 11.84AYY100 pKa = 10.23DD101 pKa = 3.37RR102 pKa = 11.84ALSDD106 pKa = 3.43EE107 pKa = 4.48KK108 pKa = 11.06KK109 pKa = 10.79ALAQQNIARR118 pKa = 11.84WRR120 pKa = 11.84DD121 pKa = 3.25FRR123 pKa = 11.84VDD125 pKa = 3.12SGQTGLIGPAGPEE138 pKa = 3.98GLSPLGYY145 pKa = 10.46KK146 pKa = 9.19GTTTFADD153 pKa = 3.7FDD155 pKa = 4.01KK156 pKa = 11.55GMFAYY161 pKa = 9.15STVEE165 pKa = 3.98NLDD168 pKa = 3.33NGNNMTFTFGDD179 pKa = 3.31GTMTQFSMPEE189 pKa = 3.75QLSLSRR195 pKa = 11.84NEE197 pKa = 4.16SASPEE202 pKa = 4.18TVITQMPYY210 pKa = 9.74NQLNAEE216 pKa = 4.53GEE218 pKa = 4.17DD219 pKa = 3.32ADD221 pKa = 4.2YY222 pKa = 11.6QEE224 pKa = 4.86LQANGSEE231 pKa = 4.22PPYY234 pKa = 10.81DD235 pKa = 3.96ADD237 pKa = 5.25AFPKK241 pKa = 10.29YY242 pKa = 9.84IWTKK246 pKa = 10.65VGTLGPFGTAIGAGGTYY263 pKa = 10.26DD264 pKa = 3.47KK265 pKa = 11.21QIVYY269 pKa = 10.48QSTTGYY275 pKa = 10.59FDD277 pKa = 4.28APCGMVLIEE286 pKa = 4.08NHH288 pKa = 6.0LTDD291 pKa = 4.06GDD293 pKa = 4.07ATVTTTIDD301 pKa = 3.4VEE303 pKa = 4.61VQAGNYY309 pKa = 9.48LGVHH313 pKa = 6.46ASVMGG318 pKa = 3.67
MM1 pKa = 7.37NMAKK5 pKa = 8.92RR6 pKa = 11.84TNKK9 pKa = 8.16THH11 pKa = 6.14AQRR14 pKa = 11.84AFQIDD19 pKa = 4.16TIGGSGQKK27 pKa = 9.76QLIQLDD33 pKa = 3.71ACLSKK38 pKa = 10.91LNHH41 pKa = 6.1RR42 pKa = 11.84LYY44 pKa = 10.74RR45 pKa = 11.84QGRR48 pKa = 11.84MYY50 pKa = 10.25RR51 pKa = 11.84AKK53 pKa = 10.76VNLPASQMPVVNSEE67 pKa = 4.51PFPQFPQTKK76 pKa = 9.71GIEE79 pKa = 3.98VWALMDD85 pKa = 3.43TWYY88 pKa = 10.73VHH90 pKa = 6.82AMWKK94 pKa = 7.54QAKK97 pKa = 9.06RR98 pKa = 11.84AYY100 pKa = 10.23DD101 pKa = 3.37RR102 pKa = 11.84ALSDD106 pKa = 3.43EE107 pKa = 4.48KK108 pKa = 11.06KK109 pKa = 10.79ALAQQNIARR118 pKa = 11.84WRR120 pKa = 11.84DD121 pKa = 3.25FRR123 pKa = 11.84VDD125 pKa = 3.12SGQTGLIGPAGPEE138 pKa = 3.98GLSPLGYY145 pKa = 10.46KK146 pKa = 9.19GTTTFADD153 pKa = 3.7FDD155 pKa = 4.01KK156 pKa = 11.55GMFAYY161 pKa = 9.15STVEE165 pKa = 3.98NLDD168 pKa = 3.33NGNNMTFTFGDD179 pKa = 3.31GTMTQFSMPEE189 pKa = 3.75QLSLSRR195 pKa = 11.84NEE197 pKa = 4.16SASPEE202 pKa = 4.18TVITQMPYY210 pKa = 9.74NQLNAEE216 pKa = 4.53GEE218 pKa = 4.17DD219 pKa = 3.32ADD221 pKa = 4.2YY222 pKa = 11.6QEE224 pKa = 4.86LQANGSEE231 pKa = 4.22PPYY234 pKa = 10.81DD235 pKa = 3.96ADD237 pKa = 5.25AFPKK241 pKa = 10.29YY242 pKa = 9.84IWTKK246 pKa = 10.65VGTLGPFGTAIGAGGTYY263 pKa = 10.26DD264 pKa = 3.47KK265 pKa = 11.21QIVYY269 pKa = 10.48QSTTGYY275 pKa = 10.59FDD277 pKa = 4.28APCGMVLIEE286 pKa = 4.08NHH288 pKa = 6.0LTDD291 pKa = 4.06GDD293 pKa = 4.07ATVTTTIDD301 pKa = 3.4VEE303 pKa = 4.61VQAGNYY309 pKa = 9.48LGVHH313 pKa = 6.46ASVMGG318 pKa = 3.67
Molecular weight: 35.07 kDa
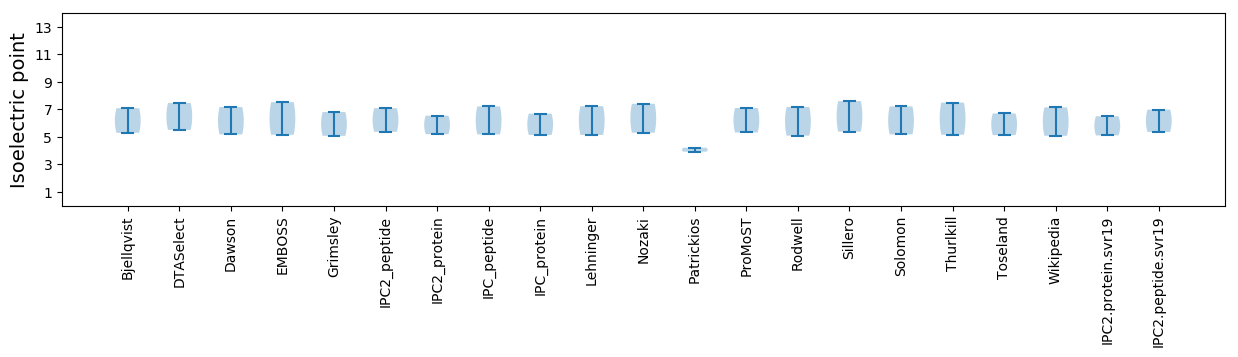
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1RKT4|M1RKT4_9VIRU Putative capsid protein OS=Gastropod associated circular ssDNA virus OX=1296573 PE=4 SV=1
MM1 pKa = 6.98AQRR4 pKa = 11.84VRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 10.25CLTLNNPTPEE18 pKa = 4.22EE19 pKa = 4.04CHH21 pKa = 6.42HH22 pKa = 6.69LLTLRR27 pKa = 11.84EE28 pKa = 4.33SVFKK32 pKa = 10.81RR33 pKa = 11.84GFFASEE39 pKa = 4.03VGKK42 pKa = 10.73KK43 pKa = 7.82GTPHH47 pKa = 6.08LQGFVHH53 pKa = 6.87LKK55 pKa = 8.94NAKK58 pKa = 8.4TLTGLKK64 pKa = 10.03KK65 pKa = 10.45FLGSDD70 pKa = 3.77RR71 pKa = 11.84YY72 pKa = 9.85HH73 pKa = 6.47VKK75 pKa = 9.84QANGTDD81 pKa = 3.71YY82 pKa = 11.48EE83 pKa = 4.08NWQYY87 pKa = 11.3IGLLNEE93 pKa = 4.83GKK95 pKa = 10.21AQGDD99 pKa = 4.73LIVQWGEE106 pKa = 3.99TPTEE110 pKa = 4.2EE111 pKa = 4.87GDD113 pKa = 3.26PDD115 pKa = 3.32AWDD118 pKa = 4.73SILEE122 pKa = 4.16MIEE125 pKa = 4.29GGFNNRR131 pKa = 11.84DD132 pKa = 3.35IVRR135 pKa = 11.84KK136 pKa = 8.01WPSIAIRR143 pKa = 11.84CQSAIDD149 pKa = 3.92RR150 pKa = 11.84YY151 pKa = 9.52RR152 pKa = 11.84VEE154 pKa = 4.39YY155 pKa = 10.33EE156 pKa = 3.49WGEE159 pKa = 3.93CRR161 pKa = 11.84AWRR164 pKa = 11.84DD165 pKa = 3.32VEE167 pKa = 4.07VEE169 pKa = 4.3YY170 pKa = 10.77IGGPTGTGKK179 pKa = 8.23TRR181 pKa = 11.84GVLYY185 pKa = 10.55HH186 pKa = 7.14ADD188 pKa = 3.1GRR190 pKa = 11.84VNTDD194 pKa = 2.31VYY196 pKa = 11.0RR197 pKa = 11.84CTNGKK202 pKa = 9.14HH203 pKa = 6.37PFDD206 pKa = 4.9KK207 pKa = 11.12YY208 pKa = 10.87DD209 pKa = 3.63GEE211 pKa = 4.34GTIVFEE217 pKa = 4.61EE218 pKa = 4.53FRR220 pKa = 11.84SQYY223 pKa = 7.73TCRR226 pKa = 11.84DD227 pKa = 3.34MLNWIDD233 pKa = 3.89GHH235 pKa = 6.51PLLLPARR242 pKa = 11.84YY243 pKa = 9.19ADD245 pKa = 3.78RR246 pKa = 11.84MAKK249 pKa = 7.73FTKK252 pKa = 10.51VIILSNWRR260 pKa = 11.84FAEE263 pKa = 4.02QYY265 pKa = 8.51RR266 pKa = 11.84TVAEE270 pKa = 4.91DD271 pKa = 3.33SPEE274 pKa = 4.11TYY276 pKa = 10.11KK277 pKa = 10.99AWLRR281 pKa = 11.84RR282 pKa = 11.84VGTITEE288 pKa = 3.9QWVV291 pKa = 3.03
MM1 pKa = 6.98AQRR4 pKa = 11.84VRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 10.25CLTLNNPTPEE18 pKa = 4.22EE19 pKa = 4.04CHH21 pKa = 6.42HH22 pKa = 6.69LLTLRR27 pKa = 11.84EE28 pKa = 4.33SVFKK32 pKa = 10.81RR33 pKa = 11.84GFFASEE39 pKa = 4.03VGKK42 pKa = 10.73KK43 pKa = 7.82GTPHH47 pKa = 6.08LQGFVHH53 pKa = 6.87LKK55 pKa = 8.94NAKK58 pKa = 8.4TLTGLKK64 pKa = 10.03KK65 pKa = 10.45FLGSDD70 pKa = 3.77RR71 pKa = 11.84YY72 pKa = 9.85HH73 pKa = 6.47VKK75 pKa = 9.84QANGTDD81 pKa = 3.71YY82 pKa = 11.48EE83 pKa = 4.08NWQYY87 pKa = 11.3IGLLNEE93 pKa = 4.83GKK95 pKa = 10.21AQGDD99 pKa = 4.73LIVQWGEE106 pKa = 3.99TPTEE110 pKa = 4.2EE111 pKa = 4.87GDD113 pKa = 3.26PDD115 pKa = 3.32AWDD118 pKa = 4.73SILEE122 pKa = 4.16MIEE125 pKa = 4.29GGFNNRR131 pKa = 11.84DD132 pKa = 3.35IVRR135 pKa = 11.84KK136 pKa = 8.01WPSIAIRR143 pKa = 11.84CQSAIDD149 pKa = 3.92RR150 pKa = 11.84YY151 pKa = 9.52RR152 pKa = 11.84VEE154 pKa = 4.39YY155 pKa = 10.33EE156 pKa = 3.49WGEE159 pKa = 3.93CRR161 pKa = 11.84AWRR164 pKa = 11.84DD165 pKa = 3.32VEE167 pKa = 4.07VEE169 pKa = 4.3YY170 pKa = 10.77IGGPTGTGKK179 pKa = 8.23TRR181 pKa = 11.84GVLYY185 pKa = 10.55HH186 pKa = 7.14ADD188 pKa = 3.1GRR190 pKa = 11.84VNTDD194 pKa = 2.31VYY196 pKa = 11.0RR197 pKa = 11.84CTNGKK202 pKa = 9.14HH203 pKa = 6.37PFDD206 pKa = 4.9KK207 pKa = 11.12YY208 pKa = 10.87DD209 pKa = 3.63GEE211 pKa = 4.34GTIVFEE217 pKa = 4.61EE218 pKa = 4.53FRR220 pKa = 11.84SQYY223 pKa = 7.73TCRR226 pKa = 11.84DD227 pKa = 3.34MLNWIDD233 pKa = 3.89GHH235 pKa = 6.51PLLLPARR242 pKa = 11.84YY243 pKa = 9.19ADD245 pKa = 3.78RR246 pKa = 11.84MAKK249 pKa = 7.73FTKK252 pKa = 10.51VIILSNWRR260 pKa = 11.84FAEE263 pKa = 4.02QYY265 pKa = 8.51RR266 pKa = 11.84TVAEE270 pKa = 4.91DD271 pKa = 3.33SPEE274 pKa = 4.11TYY276 pKa = 10.11KK277 pKa = 10.99AWLRR281 pKa = 11.84RR282 pKa = 11.84VGTITEE288 pKa = 3.9QWVV291 pKa = 3.03
Molecular weight: 33.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
609 |
291 |
318 |
304.5 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.389 ± 1.242 | 1.314 ± 0.491 |
6.076 ± 0.153 | 6.076 ± 1.201 |
3.941 ± 0.106 | 9.195 ± 0.171 |
2.135 ± 0.404 | 4.269 ± 0.356 |
5.09 ± 0.268 | 6.897 ± 0.21 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.791 ± 0.931 | 4.598 ± 0.311 |
4.433 ± 0.429 | 5.255 ± 1.194 |
5.911 ± 1.534 | 4.105 ± 0.665 |
7.882 ± 0.437 | 5.583 ± 0.396 |
2.463 ± 0.639 | 4.598 ± 0.14 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
