
Nitrobacter winogradskyi (strain ATCC 25391 / DSM 10237 / CIP 104748 / NCIMB 11846 / Nb-255)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Nitrobacter; Nitrobacter winogradskyi
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
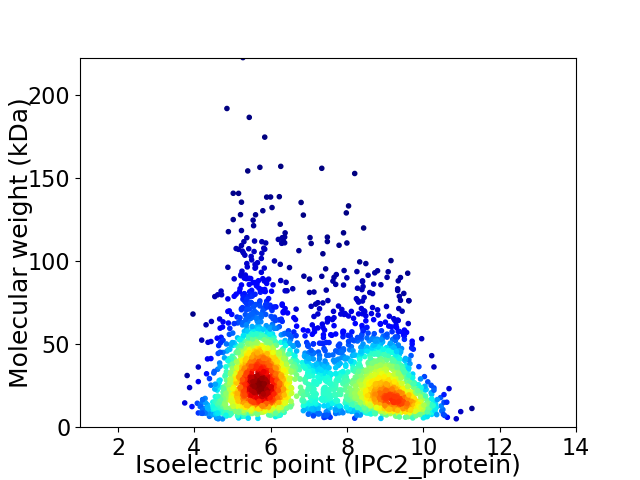
Virtual 2D-PAGE plot for 2980 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q3SU20|Q3SU20_NITWN Uncharacterized protein OS=Nitrobacter winogradskyi (strain ATCC 25391 / DSM 10237 / CIP 104748 / NCIMB 11846 / Nb-255) OX=323098 GN=Nwi_0959 PE=4 SV=1
MM1 pKa = 7.51ISGGITEE8 pKa = 5.3IIWHH12 pKa = 5.79FAGYY16 pKa = 10.4LRR18 pKa = 11.84IFDD21 pKa = 5.01DD22 pKa = 3.21IARR25 pKa = 11.84FRR27 pKa = 11.84IAHH30 pKa = 6.83DD31 pKa = 3.29EE32 pKa = 3.93GAYY35 pKa = 9.99RR36 pKa = 11.84SHH38 pKa = 6.64TEE40 pKa = 3.93DD41 pKa = 3.35YY42 pKa = 7.6TTNRR46 pKa = 11.84PQFAFTPIDD55 pKa = 3.75DD56 pKa = 4.32EE57 pKa = 4.36MSSRR61 pKa = 11.84PVAIQNPSVMGGAGHH76 pKa = 7.26HH77 pKa = 6.79APTSHH82 pKa = 7.3LSPVPDD88 pKa = 3.64VGPDD92 pKa = 3.41KK93 pKa = 10.74INPIHH98 pKa = 7.13ASGFLPPMSPGAGGGVGGSDD118 pKa = 3.84VIKK121 pKa = 10.67VVYY124 pKa = 9.9QEE126 pKa = 4.78EE127 pKa = 4.97GAQSEE132 pKa = 4.68VQTRR136 pKa = 11.84QFNHH140 pKa = 6.39MYY142 pKa = 11.05DD143 pKa = 3.32NDD145 pKa = 4.32ALVGTQDD152 pKa = 3.5SNLLIALRR160 pKa = 11.84AEE162 pKa = 4.1SDD164 pKa = 3.35HH165 pKa = 7.19AIATMIDD172 pKa = 3.76GADD175 pKa = 3.34SAIPVDD181 pKa = 3.32WRR183 pKa = 11.84VPSNTADD190 pKa = 3.34LPAFVVKK197 pKa = 10.09HH198 pKa = 5.3HH199 pKa = 6.17QALADD204 pKa = 4.09RR205 pKa = 11.84DD206 pKa = 4.16GAPDD210 pKa = 3.58PHH212 pKa = 7.87SVQQGYY218 pKa = 8.26YY219 pKa = 9.98VNGVLQDD226 pKa = 3.64PALRR230 pKa = 11.84PPDD233 pKa = 3.6QLSASDD239 pKa = 4.21PAPVPDD245 pKa = 5.05FGSGPGQWALDD256 pKa = 3.88GGNTATNGAYY266 pKa = 9.39IVDD269 pKa = 4.16LTGSARR275 pKa = 11.84TMLVVGDD282 pKa = 4.0YY283 pKa = 10.67YY284 pKa = 10.34KK285 pKa = 11.04TNAIVQTNSYY295 pKa = 9.02MDD297 pKa = 4.06NDD299 pKa = 3.94HH300 pKa = 5.82VTVGGSGAGITTGGNQATNVADD322 pKa = 4.81FVQNPGVYY330 pKa = 10.14EE331 pKa = 4.26SMPSSFAGPNWSVDD345 pKa = 3.73VVKK348 pKa = 10.87GDD350 pKa = 3.61YY351 pKa = 10.71FNIRR355 pKa = 11.84ILLQSNYY362 pKa = 10.45LSDD365 pKa = 4.35NDD367 pKa = 3.39VTMQSSSSTHH377 pKa = 5.65YY378 pKa = 10.06EE379 pKa = 3.44VHH381 pKa = 6.68AGQNEE386 pKa = 4.24LEE388 pKa = 4.24NFVLINDD395 pKa = 3.71GNFNYY400 pKa = 10.5DD401 pKa = 3.54VIVVGGSYY409 pKa = 10.49HH410 pKa = 5.73GMNVIFQNNILYY422 pKa = 10.22NNDD425 pKa = 2.97MVMMTGDD432 pKa = 3.68GVEE435 pKa = 4.33PGQTVSTGGNQLTNTATIEE454 pKa = 4.36SYY456 pKa = 11.07GGADD460 pKa = 3.49TQPWNSDD467 pKa = 3.29LDD469 pKa = 4.15SLIHH473 pKa = 6.38NISGGATEE481 pKa = 5.28LDD483 pKa = 3.56PSFGEE488 pKa = 4.56LIAGNGGTLNILYY501 pKa = 8.72ITGDD505 pKa = 3.91YY506 pKa = 10.92YY507 pKa = 11.35DD508 pKa = 5.48VNAIWQTNIVADD520 pKa = 4.02ANMAIQLQGSPSEE533 pKa = 4.34VTAGYY538 pKa = 10.26YY539 pKa = 9.96GDD541 pKa = 5.23SMTTQNVATGGNVLSNDD558 pKa = 3.23AVIVDD563 pKa = 3.74VGSTNIHH570 pKa = 6.06VNGQVYY576 pKa = 10.47GDD578 pKa = 4.65SILIQANLVEE588 pKa = 4.51EE589 pKa = 5.59DD590 pKa = 3.78SDD592 pKa = 4.64QVTQTDD598 pKa = 3.75PQALVPEE605 pKa = 4.89VIAFIGPNDD614 pKa = 3.89TNDD617 pKa = 3.53NSDD620 pKa = 3.97TQPHH624 pKa = 5.99ATTSIPHH631 pKa = 7.69DD632 pKa = 3.9DD633 pKa = 3.45PMANILHH640 pKa = 6.58
MM1 pKa = 7.51ISGGITEE8 pKa = 5.3IIWHH12 pKa = 5.79FAGYY16 pKa = 10.4LRR18 pKa = 11.84IFDD21 pKa = 5.01DD22 pKa = 3.21IARR25 pKa = 11.84FRR27 pKa = 11.84IAHH30 pKa = 6.83DD31 pKa = 3.29EE32 pKa = 3.93GAYY35 pKa = 9.99RR36 pKa = 11.84SHH38 pKa = 6.64TEE40 pKa = 3.93DD41 pKa = 3.35YY42 pKa = 7.6TTNRR46 pKa = 11.84PQFAFTPIDD55 pKa = 3.75DD56 pKa = 4.32EE57 pKa = 4.36MSSRR61 pKa = 11.84PVAIQNPSVMGGAGHH76 pKa = 7.26HH77 pKa = 6.79APTSHH82 pKa = 7.3LSPVPDD88 pKa = 3.64VGPDD92 pKa = 3.41KK93 pKa = 10.74INPIHH98 pKa = 7.13ASGFLPPMSPGAGGGVGGSDD118 pKa = 3.84VIKK121 pKa = 10.67VVYY124 pKa = 9.9QEE126 pKa = 4.78EE127 pKa = 4.97GAQSEE132 pKa = 4.68VQTRR136 pKa = 11.84QFNHH140 pKa = 6.39MYY142 pKa = 11.05DD143 pKa = 3.32NDD145 pKa = 4.32ALVGTQDD152 pKa = 3.5SNLLIALRR160 pKa = 11.84AEE162 pKa = 4.1SDD164 pKa = 3.35HH165 pKa = 7.19AIATMIDD172 pKa = 3.76GADD175 pKa = 3.34SAIPVDD181 pKa = 3.32WRR183 pKa = 11.84VPSNTADD190 pKa = 3.34LPAFVVKK197 pKa = 10.09HH198 pKa = 5.3HH199 pKa = 6.17QALADD204 pKa = 4.09RR205 pKa = 11.84DD206 pKa = 4.16GAPDD210 pKa = 3.58PHH212 pKa = 7.87SVQQGYY218 pKa = 8.26YY219 pKa = 9.98VNGVLQDD226 pKa = 3.64PALRR230 pKa = 11.84PPDD233 pKa = 3.6QLSASDD239 pKa = 4.21PAPVPDD245 pKa = 5.05FGSGPGQWALDD256 pKa = 3.88GGNTATNGAYY266 pKa = 9.39IVDD269 pKa = 4.16LTGSARR275 pKa = 11.84TMLVVGDD282 pKa = 4.0YY283 pKa = 10.67YY284 pKa = 10.34KK285 pKa = 11.04TNAIVQTNSYY295 pKa = 9.02MDD297 pKa = 4.06NDD299 pKa = 3.94HH300 pKa = 5.82VTVGGSGAGITTGGNQATNVADD322 pKa = 4.81FVQNPGVYY330 pKa = 10.14EE331 pKa = 4.26SMPSSFAGPNWSVDD345 pKa = 3.73VVKK348 pKa = 10.87GDD350 pKa = 3.61YY351 pKa = 10.71FNIRR355 pKa = 11.84ILLQSNYY362 pKa = 10.45LSDD365 pKa = 4.35NDD367 pKa = 3.39VTMQSSSSTHH377 pKa = 5.65YY378 pKa = 10.06EE379 pKa = 3.44VHH381 pKa = 6.68AGQNEE386 pKa = 4.24LEE388 pKa = 4.24NFVLINDD395 pKa = 3.71GNFNYY400 pKa = 10.5DD401 pKa = 3.54VIVVGGSYY409 pKa = 10.49HH410 pKa = 5.73GMNVIFQNNILYY422 pKa = 10.22NNDD425 pKa = 2.97MVMMTGDD432 pKa = 3.68GVEE435 pKa = 4.33PGQTVSTGGNQLTNTATIEE454 pKa = 4.36SYY456 pKa = 11.07GGADD460 pKa = 3.49TQPWNSDD467 pKa = 3.29LDD469 pKa = 4.15SLIHH473 pKa = 6.38NISGGATEE481 pKa = 5.28LDD483 pKa = 3.56PSFGEE488 pKa = 4.56LIAGNGGTLNILYY501 pKa = 8.72ITGDD505 pKa = 3.91YY506 pKa = 10.92YY507 pKa = 11.35DD508 pKa = 5.48VNAIWQTNIVADD520 pKa = 4.02ANMAIQLQGSPSEE533 pKa = 4.34VTAGYY538 pKa = 10.26YY539 pKa = 9.96GDD541 pKa = 5.23SMTTQNVATGGNVLSNDD558 pKa = 3.23AVIVDD563 pKa = 3.74VGSTNIHH570 pKa = 6.06VNGQVYY576 pKa = 10.47GDD578 pKa = 4.65SILIQANLVEE588 pKa = 4.51EE589 pKa = 5.59DD590 pKa = 3.78SDD592 pKa = 4.64QVTQTDD598 pKa = 3.75PQALVPEE605 pKa = 4.89VIAFIGPNDD614 pKa = 3.89TNDD617 pKa = 3.53NSDD620 pKa = 3.97TQPHH624 pKa = 5.99ATTSIPHH631 pKa = 7.69DD632 pKa = 3.9DD633 pKa = 3.45PMANILHH640 pKa = 6.58
Molecular weight: 68.1 kDa
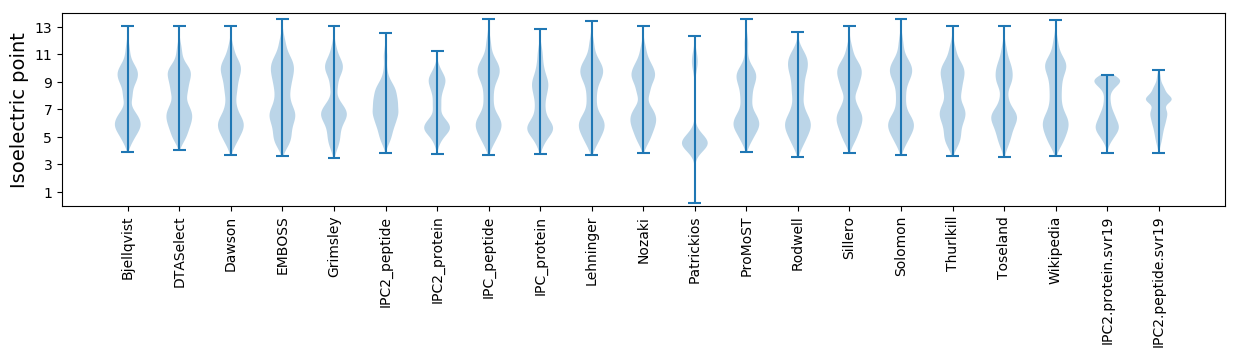
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q3SP89|Q3SP89_NITWN Cytochrome c class IC OS=Nitrobacter winogradskyi (strain ATCC 25391 / DSM 10237 / CIP 104748 / NCIMB 11846 / Nb-255) OX=323098 GN=Nwi_2649 PE=4 SV=1
MM1 pKa = 7.6LLKK4 pKa = 10.59KK5 pKa = 10.27ILGFAALGALLFFAAPAQQAQAATPVSPGIVAGQSGAGKK44 pKa = 8.85LTTNVQWGHH53 pKa = 4.43HH54 pKa = 5.13HH55 pKa = 6.0HH56 pKa = 6.25RR57 pKa = 11.84HH58 pKa = 4.67WRR60 pKa = 11.84HH61 pKa = 4.36QRR63 pKa = 11.84HH64 pKa = 5.86HH65 pKa = 6.13RR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 5.08WRR70 pKa = 11.84NHH72 pKa = 3.53RR73 pKa = 11.84HH74 pKa = 6.13HH75 pKa = 6.26RR76 pKa = 11.84HH77 pKa = 3.84NWNRR81 pKa = 11.84HH82 pKa = 3.67HH83 pKa = 6.76HH84 pKa = 5.65RR85 pKa = 11.84HH86 pKa = 5.23HH87 pKa = 5.99HH88 pKa = 5.19HH89 pKa = 6.09HH90 pKa = 5.82RR91 pKa = 11.84HH92 pKa = 4.02WRR94 pKa = 3.45
MM1 pKa = 7.6LLKK4 pKa = 10.59KK5 pKa = 10.27ILGFAALGALLFFAAPAQQAQAATPVSPGIVAGQSGAGKK44 pKa = 8.85LTTNVQWGHH53 pKa = 4.43HH54 pKa = 5.13HH55 pKa = 6.0HH56 pKa = 6.25RR57 pKa = 11.84HH58 pKa = 4.67WRR60 pKa = 11.84HH61 pKa = 4.36QRR63 pKa = 11.84HH64 pKa = 5.86HH65 pKa = 6.13RR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 5.08WRR70 pKa = 11.84NHH72 pKa = 3.53RR73 pKa = 11.84HH74 pKa = 6.13HH75 pKa = 6.26RR76 pKa = 11.84HH77 pKa = 3.84NWNRR81 pKa = 11.84HH82 pKa = 3.67HH83 pKa = 6.76HH84 pKa = 5.65RR85 pKa = 11.84HH86 pKa = 5.23HH87 pKa = 5.99HH88 pKa = 5.19HH89 pKa = 6.09HH90 pKa = 5.82RR91 pKa = 11.84HH92 pKa = 4.02WRR94 pKa = 3.45
Molecular weight: 11.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923695 |
41 |
2002 |
310.0 |
33.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.204 ± 0.067 | 0.834 ± 0.015 |
5.876 ± 0.032 | 5.489 ± 0.039 |
3.659 ± 0.031 | 8.203 ± 0.046 |
2.062 ± 0.022 | 5.323 ± 0.031 |
3.57 ± 0.035 | 9.565 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.367 ± 0.021 | 2.771 ± 0.027 |
5.294 ± 0.033 | 3.121 ± 0.027 |
7.868 ± 0.047 | 5.761 ± 0.033 |
5.348 ± 0.033 | 7.243 ± 0.035 |
1.275 ± 0.019 | 2.167 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
