
human papillomavirus 166
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 19
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
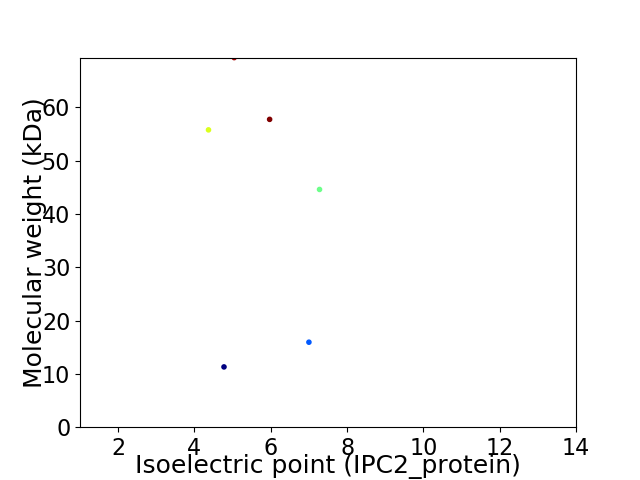
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MND3|K4MND3_9PAPI Protein E7 OS=human papillomavirus 166 OX=1315259 GN=E7 PE=3 SV=1
MM1 pKa = 6.92YY2 pKa = 10.03RR3 pKa = 11.84AGRR6 pKa = 11.84NKK8 pKa = 9.79RR9 pKa = 11.84ASAEE13 pKa = 3.94EE14 pKa = 4.76LYY16 pKa = 10.31KK17 pKa = 10.67QCATGDD23 pKa = 3.84CPPDD27 pKa = 3.37VKK29 pKa = 11.18NKK31 pKa = 10.16IEE33 pKa = 4.24GDD35 pKa = 3.2TWADD39 pKa = 3.91RR40 pKa = 11.84LLKK43 pKa = 10.08WFGSVVYY50 pKa = 9.95FGGLGIGTGRR60 pKa = 11.84GSGGSTGYY68 pKa = 10.41RR69 pKa = 11.84PLGAGTSRR77 pKa = 11.84PIGEE81 pKa = 5.01SIPIRR86 pKa = 11.84PAVPLDD92 pKa = 4.17PIGPAEE98 pKa = 4.55ILPIDD103 pKa = 3.95TANPAGPAIVQLTDD117 pKa = 2.99VTLPDD122 pKa = 4.18PSIIDD127 pKa = 3.17ISNPTTEE134 pKa = 4.67LGAGEE139 pKa = 4.3IDD141 pKa = 3.22IVSAIDD147 pKa = 3.57PLTDD151 pKa = 2.9IGGVGGNPTIISSTDD166 pKa = 2.81EE167 pKa = 4.14SAVLDD172 pKa = 4.12VQPIPPPKK180 pKa = 10.1RR181 pKa = 11.84FALDD185 pKa = 3.24VGNKK189 pKa = 9.62PSGTHH194 pKa = 4.57LTVYY198 pKa = 10.5SATTHH203 pKa = 7.24PDD205 pKa = 2.88PDD207 pKa = 3.35INIFVTSGFEE217 pKa = 4.02GEE219 pKa = 4.14IVGDD223 pKa = 3.7VEE225 pKa = 4.82EE226 pKa = 5.13IPLEE230 pKa = 4.28TFNTMQEE237 pKa = 4.31FEE239 pKa = 4.43LQEE242 pKa = 4.18PAQKK246 pKa = 9.78TSTPSDD252 pKa = 3.35VLTRR256 pKa = 11.84FVGRR260 pKa = 11.84ARR262 pKa = 11.84NLYY265 pKa = 10.31NRR267 pKa = 11.84FTEE270 pKa = 4.34QVPTQNPYY278 pKa = 10.88FLGQASRR285 pKa = 11.84AVQFEE290 pKa = 4.18FEE292 pKa = 4.33NPAFEE297 pKa = 5.8DD298 pKa = 3.76DD299 pKa = 3.66VSFTFEE305 pKa = 3.92RR306 pKa = 11.84DD307 pKa = 2.96IAEE310 pKa = 4.06IQAAPDD316 pKa = 3.24ADD318 pKa = 3.55FRR320 pKa = 11.84DD321 pKa = 3.3IRR323 pKa = 11.84VLHH326 pKa = 6.32RR327 pKa = 11.84PSYY330 pKa = 8.81FTTDD334 pKa = 2.62TGLIRR339 pKa = 11.84VSRR342 pKa = 11.84LGEE345 pKa = 3.71RR346 pKa = 11.84ATITTRR352 pKa = 11.84SGLEE356 pKa = 3.66LGRR359 pKa = 11.84PVHH362 pKa = 6.95FYY364 pKa = 11.16QDD366 pKa = 2.68ISTIEE371 pKa = 3.84PADD374 pKa = 3.95IIEE377 pKa = 4.42LQPVNDD383 pKa = 4.02TSHH386 pKa = 7.12ISTTVNTMLDD396 pKa = 3.38NTIINPAFDD405 pKa = 3.51SDD407 pKa = 4.69LYY409 pKa = 11.58NEE411 pKa = 5.12DD412 pKa = 3.97VLLDD416 pKa = 4.31DD417 pKa = 4.14YY418 pKa = 12.1SEE420 pKa = 4.73GFEE423 pKa = 4.15NTHH426 pKa = 6.71LLVTTTDD433 pKa = 3.37ADD435 pKa = 4.0NEE437 pKa = 4.4VLTVPTLPPGPSVRR451 pKa = 11.84VFIDD455 pKa = 3.73DD456 pKa = 3.82YY457 pKa = 11.72GSGLVVFNPIITTKK471 pKa = 9.54QQTIIPSQTEE481 pKa = 3.39PHH483 pKa = 6.87LIIDD487 pKa = 4.55LFSDD491 pKa = 4.67DD492 pKa = 5.3FYY494 pKa = 11.76LHH496 pKa = 6.88PGYY499 pKa = 10.72LKK501 pKa = 10.59RR502 pKa = 11.84KK503 pKa = 8.83RR504 pKa = 11.84KK505 pKa = 9.95RR506 pKa = 11.84SDD508 pKa = 2.6IFF510 pKa = 3.93
MM1 pKa = 6.92YY2 pKa = 10.03RR3 pKa = 11.84AGRR6 pKa = 11.84NKK8 pKa = 9.79RR9 pKa = 11.84ASAEE13 pKa = 3.94EE14 pKa = 4.76LYY16 pKa = 10.31KK17 pKa = 10.67QCATGDD23 pKa = 3.84CPPDD27 pKa = 3.37VKK29 pKa = 11.18NKK31 pKa = 10.16IEE33 pKa = 4.24GDD35 pKa = 3.2TWADD39 pKa = 3.91RR40 pKa = 11.84LLKK43 pKa = 10.08WFGSVVYY50 pKa = 9.95FGGLGIGTGRR60 pKa = 11.84GSGGSTGYY68 pKa = 10.41RR69 pKa = 11.84PLGAGTSRR77 pKa = 11.84PIGEE81 pKa = 5.01SIPIRR86 pKa = 11.84PAVPLDD92 pKa = 4.17PIGPAEE98 pKa = 4.55ILPIDD103 pKa = 3.95TANPAGPAIVQLTDD117 pKa = 2.99VTLPDD122 pKa = 4.18PSIIDD127 pKa = 3.17ISNPTTEE134 pKa = 4.67LGAGEE139 pKa = 4.3IDD141 pKa = 3.22IVSAIDD147 pKa = 3.57PLTDD151 pKa = 2.9IGGVGGNPTIISSTDD166 pKa = 2.81EE167 pKa = 4.14SAVLDD172 pKa = 4.12VQPIPPPKK180 pKa = 10.1RR181 pKa = 11.84FALDD185 pKa = 3.24VGNKK189 pKa = 9.62PSGTHH194 pKa = 4.57LTVYY198 pKa = 10.5SATTHH203 pKa = 7.24PDD205 pKa = 2.88PDD207 pKa = 3.35INIFVTSGFEE217 pKa = 4.02GEE219 pKa = 4.14IVGDD223 pKa = 3.7VEE225 pKa = 4.82EE226 pKa = 5.13IPLEE230 pKa = 4.28TFNTMQEE237 pKa = 4.31FEE239 pKa = 4.43LQEE242 pKa = 4.18PAQKK246 pKa = 9.78TSTPSDD252 pKa = 3.35VLTRR256 pKa = 11.84FVGRR260 pKa = 11.84ARR262 pKa = 11.84NLYY265 pKa = 10.31NRR267 pKa = 11.84FTEE270 pKa = 4.34QVPTQNPYY278 pKa = 10.88FLGQASRR285 pKa = 11.84AVQFEE290 pKa = 4.18FEE292 pKa = 4.33NPAFEE297 pKa = 5.8DD298 pKa = 3.76DD299 pKa = 3.66VSFTFEE305 pKa = 3.92RR306 pKa = 11.84DD307 pKa = 2.96IAEE310 pKa = 4.06IQAAPDD316 pKa = 3.24ADD318 pKa = 3.55FRR320 pKa = 11.84DD321 pKa = 3.3IRR323 pKa = 11.84VLHH326 pKa = 6.32RR327 pKa = 11.84PSYY330 pKa = 8.81FTTDD334 pKa = 2.62TGLIRR339 pKa = 11.84VSRR342 pKa = 11.84LGEE345 pKa = 3.71RR346 pKa = 11.84ATITTRR352 pKa = 11.84SGLEE356 pKa = 3.66LGRR359 pKa = 11.84PVHH362 pKa = 6.95FYY364 pKa = 11.16QDD366 pKa = 2.68ISTIEE371 pKa = 3.84PADD374 pKa = 3.95IIEE377 pKa = 4.42LQPVNDD383 pKa = 4.02TSHH386 pKa = 7.12ISTTVNTMLDD396 pKa = 3.38NTIINPAFDD405 pKa = 3.51SDD407 pKa = 4.69LYY409 pKa = 11.58NEE411 pKa = 5.12DD412 pKa = 3.97VLLDD416 pKa = 4.31DD417 pKa = 4.14YY418 pKa = 12.1SEE420 pKa = 4.73GFEE423 pKa = 4.15NTHH426 pKa = 6.71LLVTTTDD433 pKa = 3.37ADD435 pKa = 4.0NEE437 pKa = 4.4VLTVPTLPPGPSVRR451 pKa = 11.84VFIDD455 pKa = 3.73DD456 pKa = 3.82YY457 pKa = 11.72GSGLVVFNPIITTKK471 pKa = 9.54QQTIIPSQTEE481 pKa = 3.39PHH483 pKa = 6.87LIIDD487 pKa = 4.55LFSDD491 pKa = 4.67DD492 pKa = 5.3FYY494 pKa = 11.76LHH496 pKa = 6.88PGYY499 pKa = 10.72LKK501 pKa = 10.59RR502 pKa = 11.84KK503 pKa = 8.83RR504 pKa = 11.84KK505 pKa = 9.95RR506 pKa = 11.84SDD508 pKa = 2.6IFF510 pKa = 3.93
Molecular weight: 55.78 kDa
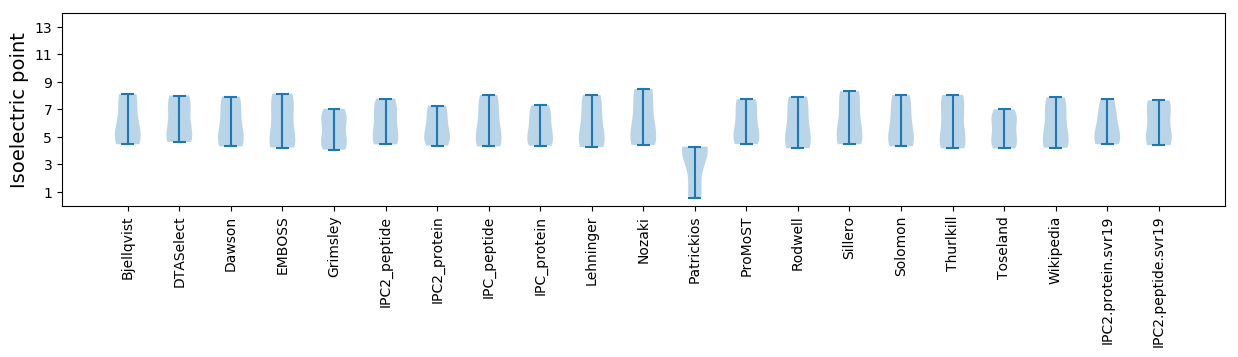
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MKR5|K4MKR5_9PAPI Major capsid protein L1 OS=human papillomavirus 166 OX=1315259 GN=L1 PE=3 SV=1
MM1 pKa = 7.89AEE3 pKa = 4.02VCPVRR8 pKa = 11.84LDD10 pKa = 3.97DD11 pKa = 3.81FCNYY15 pKa = 10.18YY16 pKa = 10.44CVSLFDD22 pKa = 5.34LSLPCIFCKK31 pKa = 10.31FICDD35 pKa = 3.97LQDD38 pKa = 3.38LASFHH43 pKa = 6.82LKK45 pKa = 10.05RR46 pKa = 11.84LSIVWRR52 pKa = 11.84SGNPFICCRR61 pKa = 11.84KK62 pKa = 9.25CVRR65 pKa = 11.84LSALYY70 pKa = 9.78EE71 pKa = 4.01AEE73 pKa = 4.22NYY75 pKa = 8.32CVCVVKK81 pKa = 10.74ASALEE86 pKa = 4.05GLLHH90 pKa = 7.25KK91 pKa = 10.19SLQSITIRR99 pKa = 11.84CLCCYY104 pKa = 10.08QLLDD108 pKa = 3.77LCEE111 pKa = 5.17KK112 pKa = 10.55LDD114 pKa = 3.78CCVRR118 pKa = 11.84DD119 pKa = 3.61EE120 pKa = 4.32NFSLVRR126 pKa = 11.84GTWRR130 pKa = 11.84GPCRR134 pKa = 11.84TCISKK139 pKa = 10.73
MM1 pKa = 7.89AEE3 pKa = 4.02VCPVRR8 pKa = 11.84LDD10 pKa = 3.97DD11 pKa = 3.81FCNYY15 pKa = 10.18YY16 pKa = 10.44CVSLFDD22 pKa = 5.34LSLPCIFCKK31 pKa = 10.31FICDD35 pKa = 3.97LQDD38 pKa = 3.38LASFHH43 pKa = 6.82LKK45 pKa = 10.05RR46 pKa = 11.84LSIVWRR52 pKa = 11.84SGNPFICCRR61 pKa = 11.84KK62 pKa = 9.25CVRR65 pKa = 11.84LSALYY70 pKa = 9.78EE71 pKa = 4.01AEE73 pKa = 4.22NYY75 pKa = 8.32CVCVVKK81 pKa = 10.74ASALEE86 pKa = 4.05GLLHH90 pKa = 7.25KK91 pKa = 10.19SLQSITIRR99 pKa = 11.84CLCCYY104 pKa = 10.08QLLDD108 pKa = 3.77LCEE111 pKa = 5.17KK112 pKa = 10.55LDD114 pKa = 3.78CCVRR118 pKa = 11.84DD119 pKa = 3.61EE120 pKa = 4.32NFSLVRR126 pKa = 11.84GTWRR130 pKa = 11.84GPCRR134 pKa = 11.84TCISKK139 pKa = 10.73
Molecular weight: 15.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2252 |
101 |
607 |
375.3 |
42.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.083 ± 0.554 | 2.709 ± 1.411 |
6.572 ± 0.644 | 6.661 ± 0.543 |
5.018 ± 0.376 | 5.551 ± 0.715 |
1.51 ± 0.315 | 4.973 ± 0.957 |
5.151 ± 0.944 | 8.659 ± 1.139 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.732 ± 0.446 | 4.796 ± 0.381 |
5.595 ± 0.931 | 4.218 ± 0.538 |
5.506 ± 0.586 | 7.194 ± 0.74 |
6.572 ± 1.032 | 6.35 ± 0.224 |
1.51 ± 0.349 | 3.641 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
