
McMurdo Ice Shelf pond-associated circular DNA virus-6
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
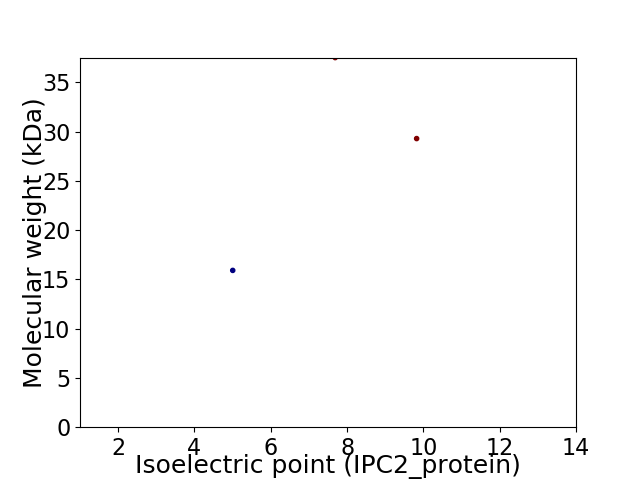
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
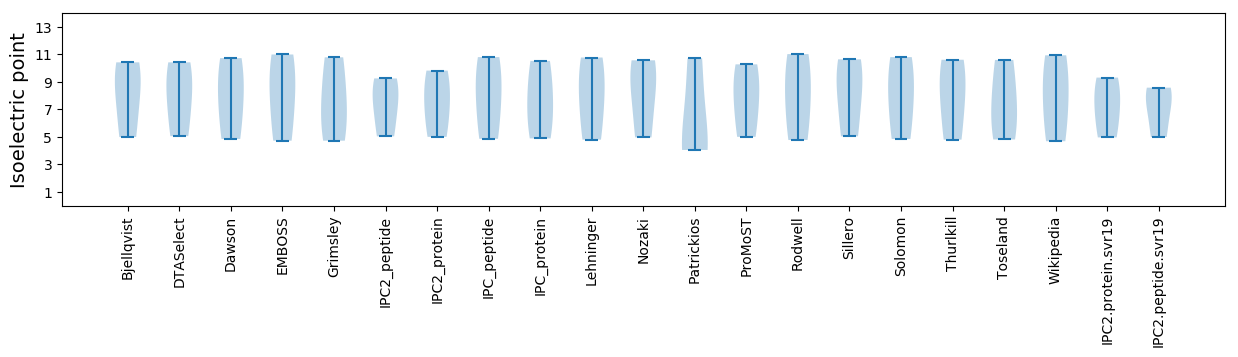
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075LZ80|A0A075LZ80_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-6 OX=1521390 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.04GQEE5 pKa = 3.79ACEE8 pKa = 4.15EE9 pKa = 4.31VLSSSGEE16 pKa = 4.1EE17 pKa = 3.82QDD19 pKa = 4.46SNPVDD24 pKa = 3.15VWDD27 pKa = 4.27EE28 pKa = 3.9VSGEE32 pKa = 4.25WKK34 pKa = 9.61HH35 pKa = 6.37HH36 pKa = 5.16PRR38 pKa = 11.84AEE40 pKa = 3.96KK41 pKa = 10.48RR42 pKa = 11.84PMGSDD47 pKa = 3.89EE48 pKa = 5.13EE49 pKa = 4.37ILDD52 pKa = 3.8NASKK56 pKa = 10.4FFQSFKK62 pKa = 10.47EE63 pKa = 3.8SLMCHH68 pKa = 5.6EE69 pKa = 5.14VWAQPDD75 pKa = 3.41LAEE78 pKa = 4.06YY79 pKa = 9.37FANFEE84 pKa = 4.06ISEE87 pKa = 4.27FTQISLCRR95 pKa = 11.84TYY97 pKa = 11.54ANYY100 pKa = 10.01LAQVMRR106 pKa = 11.84ARR108 pKa = 11.84NAGTPSEE115 pKa = 4.29GQSPPGGANSLRR127 pKa = 11.84VGHH130 pKa = 6.37TSTRR134 pKa = 11.84KK135 pKa = 9.25IKK137 pKa = 10.59RR138 pKa = 11.84KK139 pKa = 9.99LFF141 pKa = 3.81
MM1 pKa = 8.12DD2 pKa = 4.04GQEE5 pKa = 3.79ACEE8 pKa = 4.15EE9 pKa = 4.31VLSSSGEE16 pKa = 4.1EE17 pKa = 3.82QDD19 pKa = 4.46SNPVDD24 pKa = 3.15VWDD27 pKa = 4.27EE28 pKa = 3.9VSGEE32 pKa = 4.25WKK34 pKa = 9.61HH35 pKa = 6.37HH36 pKa = 5.16PRR38 pKa = 11.84AEE40 pKa = 3.96KK41 pKa = 10.48RR42 pKa = 11.84PMGSDD47 pKa = 3.89EE48 pKa = 5.13EE49 pKa = 4.37ILDD52 pKa = 3.8NASKK56 pKa = 10.4FFQSFKK62 pKa = 10.47EE63 pKa = 3.8SLMCHH68 pKa = 5.6EE69 pKa = 5.14VWAQPDD75 pKa = 3.41LAEE78 pKa = 4.06YY79 pKa = 9.37FANFEE84 pKa = 4.06ISEE87 pKa = 4.27FTQISLCRR95 pKa = 11.84TYY97 pKa = 11.54ANYY100 pKa = 10.01LAQVMRR106 pKa = 11.84ARR108 pKa = 11.84NAGTPSEE115 pKa = 4.29GQSPPGGANSLRR127 pKa = 11.84VGHH130 pKa = 6.37TSTRR134 pKa = 11.84KK135 pKa = 9.25IKK137 pKa = 10.59RR138 pKa = 11.84KK139 pKa = 9.99LFF141 pKa = 3.81
Molecular weight: 15.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075LYS7|A0A075LYS7_9VIRU ATP-dependent helicase Rep OS=McMurdo Ice Shelf pond-associated circular DNA virus-6 OX=1521390 PE=3 SV=1
MM1 pKa = 7.44SYY3 pKa = 10.5KK4 pKa = 10.44RR5 pKa = 11.84KK6 pKa = 9.37LSKK9 pKa = 9.74SARR12 pKa = 11.84YY13 pKa = 8.95NRR15 pKa = 11.84RR16 pKa = 11.84PLKK19 pKa = 10.45VISVAQKK26 pKa = 11.02AKK28 pKa = 10.02LQRR31 pKa = 11.84ARR33 pKa = 11.84SGIPKK38 pKa = 10.0VIVAASDD45 pKa = 3.34PMMKK49 pKa = 9.94QLRR52 pKa = 11.84GLLAAKK58 pKa = 9.41TRR60 pKa = 11.84DD61 pKa = 3.4ALDD64 pKa = 3.34VYY66 pKa = 11.19RR67 pKa = 11.84NATAGGGVSRR77 pKa = 11.84VSLLTSSSPFTTAAAATGILDD98 pKa = 3.83ADD100 pKa = 3.97ADD102 pKa = 4.03EE103 pKa = 4.38VLINSVRR110 pKa = 11.84IAGYY114 pKa = 10.36HH115 pKa = 6.43DD116 pKa = 3.68NQFQADD122 pKa = 4.1TDD124 pKa = 4.03PVGNFPTFVRR134 pKa = 11.84VLVVWMTKK142 pKa = 9.98PLLIASAAGTLPTVTEE158 pKa = 4.18VLTAASIDD166 pKa = 3.5AMPIQSQQNGGRR178 pKa = 11.84FTILSDD184 pKa = 3.55RR185 pKa = 11.84RR186 pKa = 11.84WNLGLATTSASVSTTSAQSMRR207 pKa = 11.84QFFDD211 pKa = 3.12YY212 pKa = 9.87TIKK215 pKa = 10.56VGRR218 pKa = 11.84RR219 pKa = 11.84CKK221 pKa = 10.24FVTPAASGAATAGGHH236 pKa = 5.91YY237 pKa = 8.75GTGAATGQLSTGLLLMYY254 pKa = 10.88SMAVGPAGLNTNVNFATRR272 pKa = 11.84VNYY275 pKa = 9.49TGG277 pKa = 3.41
MM1 pKa = 7.44SYY3 pKa = 10.5KK4 pKa = 10.44RR5 pKa = 11.84KK6 pKa = 9.37LSKK9 pKa = 9.74SARR12 pKa = 11.84YY13 pKa = 8.95NRR15 pKa = 11.84RR16 pKa = 11.84PLKK19 pKa = 10.45VISVAQKK26 pKa = 11.02AKK28 pKa = 10.02LQRR31 pKa = 11.84ARR33 pKa = 11.84SGIPKK38 pKa = 10.0VIVAASDD45 pKa = 3.34PMMKK49 pKa = 9.94QLRR52 pKa = 11.84GLLAAKK58 pKa = 9.41TRR60 pKa = 11.84DD61 pKa = 3.4ALDD64 pKa = 3.34VYY66 pKa = 11.19RR67 pKa = 11.84NATAGGGVSRR77 pKa = 11.84VSLLTSSSPFTTAAAATGILDD98 pKa = 3.83ADD100 pKa = 3.97ADD102 pKa = 4.03EE103 pKa = 4.38VLINSVRR110 pKa = 11.84IAGYY114 pKa = 10.36HH115 pKa = 6.43DD116 pKa = 3.68NQFQADD122 pKa = 4.1TDD124 pKa = 4.03PVGNFPTFVRR134 pKa = 11.84VLVVWMTKK142 pKa = 9.98PLLIASAAGTLPTVTEE158 pKa = 4.18VLTAASIDD166 pKa = 3.5AMPIQSQQNGGRR178 pKa = 11.84FTILSDD184 pKa = 3.55RR185 pKa = 11.84RR186 pKa = 11.84WNLGLATTSASVSTTSAQSMRR207 pKa = 11.84QFFDD211 pKa = 3.12YY212 pKa = 9.87TIKK215 pKa = 10.56VGRR218 pKa = 11.84RR219 pKa = 11.84CKK221 pKa = 10.24FVTPAASGAATAGGHH236 pKa = 5.91YY237 pKa = 8.75GTGAATGQLSTGLLLMYY254 pKa = 10.88SMAVGPAGLNTNVNFATRR272 pKa = 11.84VNYY275 pKa = 9.49TGG277 pKa = 3.41
Molecular weight: 29.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
744 |
141 |
326 |
248.0 |
27.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.005 ± 2.131 | 2.151 ± 0.858 |
4.704 ± 0.168 | 4.57 ± 2.443 |
5.78 ± 1.344 | 7.392 ± 0.452 |
2.957 ± 1.084 | 3.36 ± 0.283 |
5.108 ± 0.392 | 7.527 ± 0.685 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.957 ± 0.063 | 3.629 ± 0.322 |
5.511 ± 0.828 | 4.301 ± 0.231 |
6.183 ± 0.307 | 8.065 ± 1.06 |
6.317 ± 1.572 | 6.048 ± 0.843 |
1.613 ± 0.397 | 2.823 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
