
Gammarus sp. amphipod associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
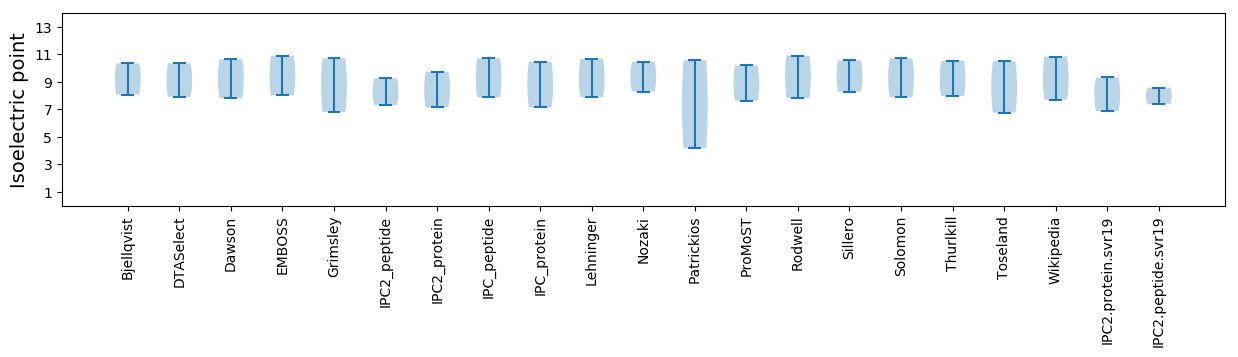
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLR1|A0A0K1RLR1_9CIRC Putative replication initiation protein OS=Gammarus sp. amphipod associated circular virus OX=1692250 PE=4 SV=1
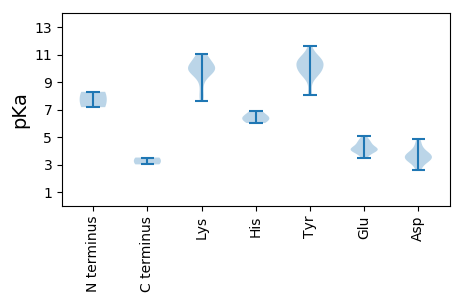
MM1 pKa = 7.17TSCRR5 pKa = 11.84YY6 pKa = 9.94CFTVNNPTVQDD17 pKa = 3.74RR18 pKa = 11.84EE19 pKa = 4.2RR20 pKa = 11.84LDD22 pKa = 4.66LLADD26 pKa = 3.44SCNYY30 pKa = 9.87LVYY33 pKa = 10.65GNEE36 pKa = 4.17IGSSGTPHH44 pKa = 6.44LQGFVIFPKK53 pKa = 9.85TKK55 pKa = 9.72RR56 pKa = 11.84FNAAKK61 pKa = 9.99IAIGNTAHH69 pKa = 6.2VEE71 pKa = 4.16CARR74 pKa = 11.84GSSVQAATYY83 pKa = 9.48CKK85 pKa = 10.0KK86 pKa = 10.81DD87 pKa = 2.88GDD89 pKa = 3.68FRR91 pKa = 11.84EE92 pKa = 4.58FGEE95 pKa = 4.77LPSSQGKK102 pKa = 7.62RR103 pKa = 11.84TDD105 pKa = 2.66WDD107 pKa = 3.33IYY109 pKa = 10.58RR110 pKa = 11.84DD111 pKa = 3.42WVTDD115 pKa = 3.88LGRR118 pKa = 11.84VPSKK122 pKa = 11.02KK123 pKa = 10.0EE124 pKa = 3.49LVLAFPGFYY133 pKa = 10.5ARR135 pKa = 11.84YY136 pKa = 9.09RR137 pKa = 11.84KK138 pKa = 10.09ACFEE142 pKa = 4.07YY143 pKa = 11.04AEE145 pKa = 4.91ALTPPPILTQSEE157 pKa = 4.61PRR159 pKa = 11.84FGWQTRR165 pKa = 11.84VDD167 pKa = 4.22GIINGEE173 pKa = 4.01ANDD176 pKa = 3.92RR177 pKa = 11.84TIHH180 pKa = 6.02FVVDD184 pKa = 3.44PEE186 pKa = 4.41GNAGKK191 pKa = 8.11TWFCSYY197 pKa = 11.61ALTKK201 pKa = 10.18WPDD204 pKa = 3.0KK205 pKa = 9.06VQVMRR210 pKa = 11.84IGKK213 pKa = 9.5RR214 pKa = 11.84DD215 pKa = 3.69DD216 pKa = 3.16LAYY219 pKa = 10.39AISTEE224 pKa = 3.86KK225 pKa = 10.77SIFLMDD231 pKa = 3.52VPRR234 pKa = 11.84NQMTFLQYY242 pKa = 11.01SVLEE246 pKa = 4.04MLKK249 pKa = 10.78DD250 pKa = 3.7RR251 pKa = 11.84MIFSPKK257 pKa = 9.59YY258 pKa = 8.08EE259 pKa = 4.15SSFKK263 pKa = 9.92ILQYY267 pKa = 9.8VPHH270 pKa = 6.86VIVFSNEE277 pKa = 3.88QPDD280 pKa = 4.11TSALSADD287 pKa = 4.82RR288 pKa = 11.84INIINVV294 pKa = 3.01
MM1 pKa = 7.17TSCRR5 pKa = 11.84YY6 pKa = 9.94CFTVNNPTVQDD17 pKa = 3.74RR18 pKa = 11.84EE19 pKa = 4.2RR20 pKa = 11.84LDD22 pKa = 4.66LLADD26 pKa = 3.44SCNYY30 pKa = 9.87LVYY33 pKa = 10.65GNEE36 pKa = 4.17IGSSGTPHH44 pKa = 6.44LQGFVIFPKK53 pKa = 9.85TKK55 pKa = 9.72RR56 pKa = 11.84FNAAKK61 pKa = 9.99IAIGNTAHH69 pKa = 6.2VEE71 pKa = 4.16CARR74 pKa = 11.84GSSVQAATYY83 pKa = 9.48CKK85 pKa = 10.0KK86 pKa = 10.81DD87 pKa = 2.88GDD89 pKa = 3.68FRR91 pKa = 11.84EE92 pKa = 4.58FGEE95 pKa = 4.77LPSSQGKK102 pKa = 7.62RR103 pKa = 11.84TDD105 pKa = 2.66WDD107 pKa = 3.33IYY109 pKa = 10.58RR110 pKa = 11.84DD111 pKa = 3.42WVTDD115 pKa = 3.88LGRR118 pKa = 11.84VPSKK122 pKa = 11.02KK123 pKa = 10.0EE124 pKa = 3.49LVLAFPGFYY133 pKa = 10.5ARR135 pKa = 11.84YY136 pKa = 9.09RR137 pKa = 11.84KK138 pKa = 10.09ACFEE142 pKa = 4.07YY143 pKa = 11.04AEE145 pKa = 4.91ALTPPPILTQSEE157 pKa = 4.61PRR159 pKa = 11.84FGWQTRR165 pKa = 11.84VDD167 pKa = 4.22GIINGEE173 pKa = 4.01ANDD176 pKa = 3.92RR177 pKa = 11.84TIHH180 pKa = 6.02FVVDD184 pKa = 3.44PEE186 pKa = 4.41GNAGKK191 pKa = 8.11TWFCSYY197 pKa = 11.61ALTKK201 pKa = 10.18WPDD204 pKa = 3.0KK205 pKa = 9.06VQVMRR210 pKa = 11.84IGKK213 pKa = 9.5RR214 pKa = 11.84DD215 pKa = 3.69DD216 pKa = 3.16LAYY219 pKa = 10.39AISTEE224 pKa = 3.86KK225 pKa = 10.77SIFLMDD231 pKa = 3.52VPRR234 pKa = 11.84NQMTFLQYY242 pKa = 11.01SVLEE246 pKa = 4.04MLKK249 pKa = 10.78DD250 pKa = 3.7RR251 pKa = 11.84MIFSPKK257 pKa = 9.59YY258 pKa = 8.08EE259 pKa = 4.15SSFKK263 pKa = 9.92ILQYY267 pKa = 9.8VPHH270 pKa = 6.86VIVFSNEE277 pKa = 3.88QPDD280 pKa = 4.11TSALSADD287 pKa = 4.82RR288 pKa = 11.84INIINVV294 pKa = 3.01
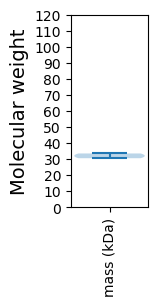
Molecular weight: 33.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLR1|A0A0K1RLR1_9CIRC Putative replication initiation protein OS=Gammarus sp. amphipod associated circular virus OX=1692250 PE=4 SV=1
MM1 pKa = 8.29PYY3 pKa = 9.65RR4 pKa = 11.84KK5 pKa = 9.45SYY7 pKa = 10.34RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TSRR13 pKa = 11.84KK14 pKa = 7.88RR15 pKa = 11.84FVRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84KK22 pKa = 9.47ARR24 pKa = 11.84AYY26 pKa = 8.12RR27 pKa = 11.84QKK29 pKa = 10.82RR30 pKa = 11.84GLDD33 pKa = 3.23KK34 pKa = 10.87TRR36 pKa = 11.84IGATPGSRR44 pKa = 11.84APCKK48 pKa = 10.33RR49 pKa = 11.84ITKK52 pKa = 9.88INQTTAQGQNDD63 pKa = 4.17RR64 pKa = 11.84EE65 pKa = 4.29LYY67 pKa = 10.37VYY69 pKa = 10.35PEE71 pKa = 4.04LTAIPKK77 pKa = 10.29GDD79 pKa = 4.49AINNRR84 pKa = 11.84QRR86 pKa = 11.84DD87 pKa = 3.43VCTIVGFRR95 pKa = 11.84INWEE99 pKa = 4.01MTVDD103 pKa = 3.66TRR105 pKa = 11.84NAMYY109 pKa = 9.84WNMAIVCPKK118 pKa = 10.22PKK120 pKa = 10.58ARR122 pKa = 11.84DD123 pKa = 3.28LAFQTGDD130 pKa = 3.31NVSTEE135 pKa = 3.74QFFRR139 pKa = 11.84GDD141 pKa = 3.24NQQRR145 pKa = 11.84GEE147 pKa = 4.31DD148 pKa = 3.81FSLSLTSQEE157 pKa = 3.89MHH159 pKa = 6.49YY160 pKa = 9.62KK161 pKa = 9.71TINSDD166 pKa = 3.77LYY168 pKa = 10.52NVLWRR173 pKa = 11.84KK174 pKa = 10.09KK175 pKa = 8.61MLINGSLPLSNEE187 pKa = 3.53EE188 pKa = 4.53QIAQNASSGKK198 pKa = 9.98NYY200 pKa = 10.47RR201 pKa = 11.84IGQKK205 pKa = 9.9YY206 pKa = 9.67IKK208 pKa = 10.17LNRR211 pKa = 11.84QMMFEE216 pKa = 5.05DD217 pKa = 3.59NQSNYY222 pKa = 10.0PIYY225 pKa = 10.74LLTWGARR232 pKa = 11.84FEE234 pKa = 4.7ADD236 pKa = 2.59SGAPISAGSPLAVRR250 pKa = 11.84MRR252 pKa = 11.84VLTFFRR258 pKa = 11.84EE259 pKa = 4.14PRR261 pKa = 11.84NN262 pKa = 3.51
MM1 pKa = 8.29PYY3 pKa = 9.65RR4 pKa = 11.84KK5 pKa = 9.45SYY7 pKa = 10.34RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TSRR13 pKa = 11.84KK14 pKa = 7.88RR15 pKa = 11.84FVRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84KK22 pKa = 9.47ARR24 pKa = 11.84AYY26 pKa = 8.12RR27 pKa = 11.84QKK29 pKa = 10.82RR30 pKa = 11.84GLDD33 pKa = 3.23KK34 pKa = 10.87TRR36 pKa = 11.84IGATPGSRR44 pKa = 11.84APCKK48 pKa = 10.33RR49 pKa = 11.84ITKK52 pKa = 9.88INQTTAQGQNDD63 pKa = 4.17RR64 pKa = 11.84EE65 pKa = 4.29LYY67 pKa = 10.37VYY69 pKa = 10.35PEE71 pKa = 4.04LTAIPKK77 pKa = 10.29GDD79 pKa = 4.49AINNRR84 pKa = 11.84QRR86 pKa = 11.84DD87 pKa = 3.43VCTIVGFRR95 pKa = 11.84INWEE99 pKa = 4.01MTVDD103 pKa = 3.66TRR105 pKa = 11.84NAMYY109 pKa = 9.84WNMAIVCPKK118 pKa = 10.22PKK120 pKa = 10.58ARR122 pKa = 11.84DD123 pKa = 3.28LAFQTGDD130 pKa = 3.31NVSTEE135 pKa = 3.74QFFRR139 pKa = 11.84GDD141 pKa = 3.24NQQRR145 pKa = 11.84GEE147 pKa = 4.31DD148 pKa = 3.81FSLSLTSQEE157 pKa = 3.89MHH159 pKa = 6.49YY160 pKa = 9.62KK161 pKa = 9.71TINSDD166 pKa = 3.77LYY168 pKa = 10.52NVLWRR173 pKa = 11.84KK174 pKa = 10.09KK175 pKa = 8.61MLINGSLPLSNEE187 pKa = 3.53EE188 pKa = 4.53QIAQNASSGKK198 pKa = 9.98NYY200 pKa = 10.47RR201 pKa = 11.84IGQKK205 pKa = 9.9YY206 pKa = 9.67IKK208 pKa = 10.17LNRR211 pKa = 11.84QMMFEE216 pKa = 5.05DD217 pKa = 3.59NQSNYY222 pKa = 10.0PIYY225 pKa = 10.74LLTWGARR232 pKa = 11.84FEE234 pKa = 4.7ADD236 pKa = 2.59SGAPISAGSPLAVRR250 pKa = 11.84MRR252 pKa = 11.84VLTFFRR258 pKa = 11.84EE259 pKa = 4.14PRR261 pKa = 11.84NN262 pKa = 3.51
Molecular weight: 30.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
556 |
262 |
294 |
278.0 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.835 ± 0.023 | 1.799 ± 0.425 |
5.576 ± 0.648 | 4.676 ± 0.311 |
4.856 ± 0.676 | 5.755 ± 0.02 |
0.899 ± 0.337 | 5.935 ± 0.137 |
5.935 ± 0.112 | 6.295 ± 0.122 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.48 | 5.755 ± 0.974 |
5.216 ± 0.165 | 4.676 ± 0.683 |
8.993 ± 1.848 | 6.475 ± 0.239 |
6.115 ± 0.005 | 5.396 ± 1.027 |
1.619 ± 0.06 | 4.496 ± 0.054 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
