
Lentinula edodes (Shiitake mushroom) (Lentinus edodes)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Omphalotaceae; Lentinula
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 12046 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
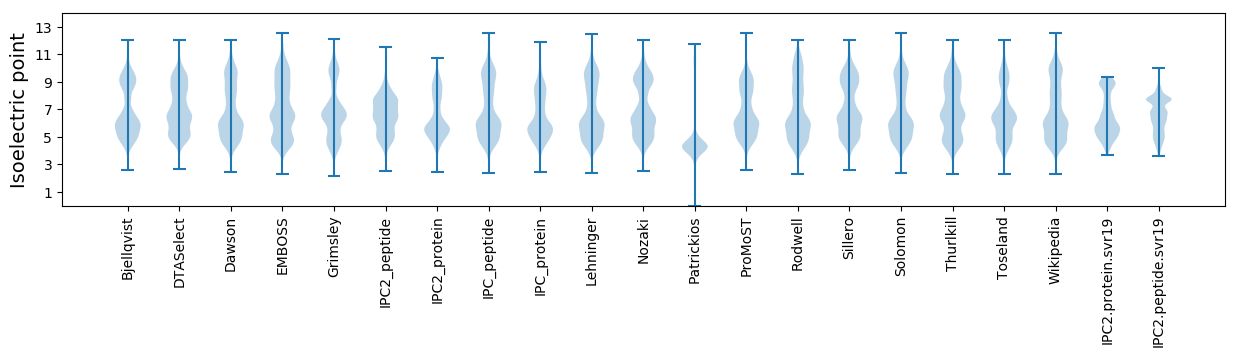
* You can choose from 21 different methods for calculating isoelectric point
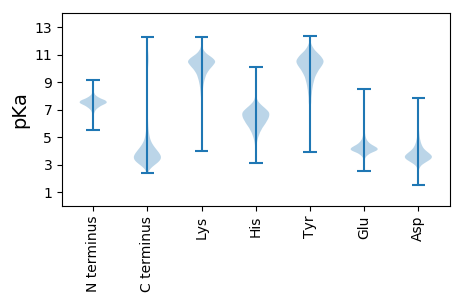
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q3E836|A0A1Q3E836_LENED MICOS complex subunit MIC10 OS=Lentinula edodes OX=5353 GN=LENED_005046 PE=3 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84SHH4 pKa = 7.28IALLSVLVYY13 pKa = 10.66SAVLEE18 pKa = 4.24ANVLKK23 pKa = 10.59SKK25 pKa = 10.64RR26 pKa = 11.84VTEE29 pKa = 4.49ASCEE33 pKa = 4.08LTASSGDD40 pKa = 3.73DD41 pKa = 3.22APQFLAAAASCDD53 pKa = 3.77TVIIPAGTTLNIEE66 pKa = 4.22TRR68 pKa = 11.84LDD70 pKa = 3.44MTGMSNLNIDD80 pKa = 3.69LQGTIRR86 pKa = 11.84FNPDD90 pKa = 2.41LDD92 pKa = 3.76YY93 pKa = 11.09WSGNGFPITFQDD105 pKa = 5.15QITFWLLGGEE115 pKa = 4.33NIVLSGGGTLDD126 pKa = 3.45GAGQAWYY133 pKa = 9.62DD134 pKa = 3.56AFASNSSLLRR144 pKa = 11.84PIILTLNEE152 pKa = 3.95ATNVLVEE159 pKa = 5.22DD160 pKa = 3.55IQMINSPEE168 pKa = 3.74WFNLVNEE175 pKa = 4.55CQNVTFNNITITAKK189 pKa = 9.39STSDD193 pKa = 3.25NFISNTDD200 pKa = 2.44GWDD203 pKa = 2.91IYY205 pKa = 10.77RR206 pKa = 11.84SDD208 pKa = 3.43MVTIKK213 pKa = 10.94NSIINNGDD221 pKa = 3.25DD222 pKa = 3.6CVSFKK227 pKa = 11.02PNSTNILVSDD237 pKa = 4.28LNCNGSHH244 pKa = 6.91GISVGSLGQFSGVFDD259 pKa = 3.52IVQNVTAVNIQMANAEE275 pKa = 3.86NGARR279 pKa = 11.84IKK281 pKa = 10.93AFAGDD286 pKa = 4.0DD287 pKa = 3.74VGSGTVQNITFQNFVVSNVDD307 pKa = 3.39SPVVIDD313 pKa = 3.33QCYY316 pKa = 7.06EE317 pKa = 3.86TSADD321 pKa = 3.67DD322 pKa = 4.57CEE324 pKa = 4.36EE325 pKa = 4.03FPSNVFIQDD334 pKa = 2.81IFFNNISGTGSGSQVASLDD353 pKa = 3.58CSPGSRR359 pKa = 11.84CTDD362 pKa = 2.84INVNGFDD369 pKa = 5.14LSGSDD374 pKa = 3.18GKK376 pKa = 7.73TTYY379 pKa = 10.18EE380 pKa = 4.11CQNVVLAGNAASLFGTCTTTT400 pKa = 3.77
MM1 pKa = 7.86RR2 pKa = 11.84SHH4 pKa = 7.28IALLSVLVYY13 pKa = 10.66SAVLEE18 pKa = 4.24ANVLKK23 pKa = 10.59SKK25 pKa = 10.64RR26 pKa = 11.84VTEE29 pKa = 4.49ASCEE33 pKa = 4.08LTASSGDD40 pKa = 3.73DD41 pKa = 3.22APQFLAAAASCDD53 pKa = 3.77TVIIPAGTTLNIEE66 pKa = 4.22TRR68 pKa = 11.84LDD70 pKa = 3.44MTGMSNLNIDD80 pKa = 3.69LQGTIRR86 pKa = 11.84FNPDD90 pKa = 2.41LDD92 pKa = 3.76YY93 pKa = 11.09WSGNGFPITFQDD105 pKa = 5.15QITFWLLGGEE115 pKa = 4.33NIVLSGGGTLDD126 pKa = 3.45GAGQAWYY133 pKa = 9.62DD134 pKa = 3.56AFASNSSLLRR144 pKa = 11.84PIILTLNEE152 pKa = 3.95ATNVLVEE159 pKa = 5.22DD160 pKa = 3.55IQMINSPEE168 pKa = 3.74WFNLVNEE175 pKa = 4.55CQNVTFNNITITAKK189 pKa = 9.39STSDD193 pKa = 3.25NFISNTDD200 pKa = 2.44GWDD203 pKa = 2.91IYY205 pKa = 10.77RR206 pKa = 11.84SDD208 pKa = 3.43MVTIKK213 pKa = 10.94NSIINNGDD221 pKa = 3.25DD222 pKa = 3.6CVSFKK227 pKa = 11.02PNSTNILVSDD237 pKa = 4.28LNCNGSHH244 pKa = 6.91GISVGSLGQFSGVFDD259 pKa = 3.52IVQNVTAVNIQMANAEE275 pKa = 3.86NGARR279 pKa = 11.84IKK281 pKa = 10.93AFAGDD286 pKa = 4.0DD287 pKa = 3.74VGSGTVQNITFQNFVVSNVDD307 pKa = 3.39SPVVIDD313 pKa = 3.33QCYY316 pKa = 7.06EE317 pKa = 3.86TSADD321 pKa = 3.67DD322 pKa = 4.57CEE324 pKa = 4.36EE325 pKa = 4.03FPSNVFIQDD334 pKa = 2.81IFFNNISGTGSGSQVASLDD353 pKa = 3.58CSPGSRR359 pKa = 11.84CTDD362 pKa = 2.84INVNGFDD369 pKa = 5.14LSGSDD374 pKa = 3.18GKK376 pKa = 7.73TTYY379 pKa = 10.18EE380 pKa = 4.11CQNVVLAGNAASLFGTCTTTT400 pKa = 3.77
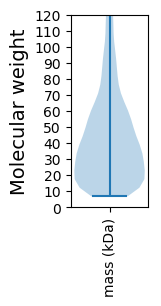
Molecular weight: 42.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q3EJC6|A0A1Q3EJC6_LENED 3'(2') 5'-bisphosphate nucleotidase OS=Lentinula edodes OX=5353 GN=LENED_009199 PE=3 SV=1
MM1 pKa = 6.88TVLWSTSGSPSSSMVAIASQQPRR24 pKa = 11.84PNTQAYY30 pKa = 9.76NNNTTNNPRR39 pKa = 11.84MILHH43 pKa = 7.14PIPRR47 pKa = 11.84LFFPAHH53 pKa = 5.78CVVDD57 pKa = 4.61RR58 pKa = 11.84SWGGSCSGEE67 pKa = 3.74RR68 pKa = 11.84GKK70 pKa = 10.47DD71 pKa = 2.87IFFQRR76 pKa = 11.84IEE78 pKa = 4.1VYY80 pKa = 10.05VEE82 pKa = 4.18KK83 pKa = 10.54KK84 pKa = 9.64RR85 pKa = 11.84KK86 pKa = 9.05RR87 pKa = 11.84PGRR90 pKa = 11.84FTLRR94 pKa = 11.84ANCHH98 pKa = 5.72ASAA101 pKa = 4.64
MM1 pKa = 6.88TVLWSTSGSPSSSMVAIASQQPRR24 pKa = 11.84PNTQAYY30 pKa = 9.76NNNTTNNPRR39 pKa = 11.84MILHH43 pKa = 7.14PIPRR47 pKa = 11.84LFFPAHH53 pKa = 5.78CVVDD57 pKa = 4.61RR58 pKa = 11.84SWGGSCSGEE67 pKa = 3.74RR68 pKa = 11.84GKK70 pKa = 10.47DD71 pKa = 2.87IFFQRR76 pKa = 11.84IEE78 pKa = 4.1VYY80 pKa = 10.05VEE82 pKa = 4.18KK83 pKa = 10.54KK84 pKa = 9.64RR85 pKa = 11.84KK86 pKa = 9.05RR87 pKa = 11.84PGRR90 pKa = 11.84FTLRR94 pKa = 11.84ANCHH98 pKa = 5.72ASAA101 pKa = 4.64
Molecular weight: 11.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5294775 |
58 |
4811 |
439.5 |
48.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.816 ± 0.019 | 1.145 ± 0.007 |
5.555 ± 0.016 | 6.044 ± 0.026 |
3.927 ± 0.014 | 6.353 ± 0.021 |
2.442 ± 0.009 | 5.332 ± 0.016 |
4.787 ± 0.023 | 9.199 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.009 | 3.957 ± 0.012 |
6.105 ± 0.024 | 3.719 ± 0.013 |
5.698 ± 0.018 | 9.293 ± 0.032 |
6.167 ± 0.016 | 6.227 ± 0.016 |
1.36 ± 0.009 | 2.785 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
