
Tobacco virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Closterovirus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
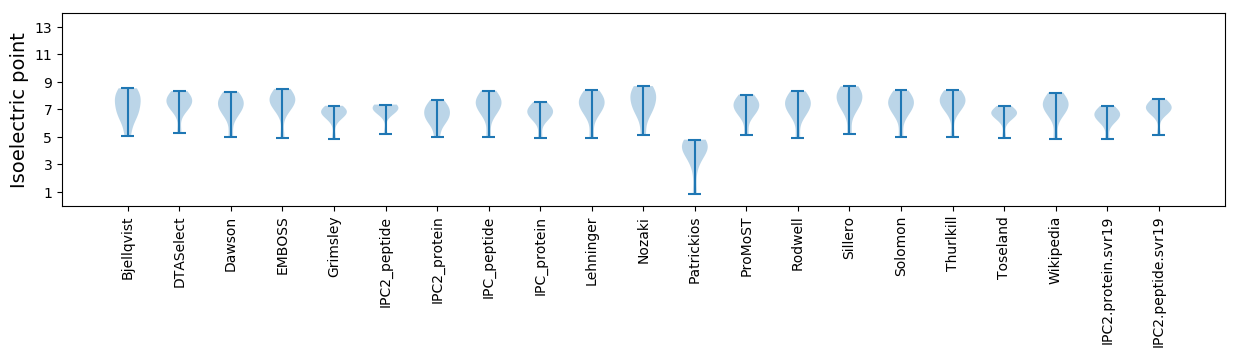
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0K1HRT1|A0A0K1HRT1_9CLOS Hsp70h OS=Tobacco virus 1 OX=1692045 GN=Hsp70h PE=4 SV=1
MM1 pKa = 7.41LSDD4 pKa = 4.21YY5 pKa = 10.96EE6 pKa = 4.37VGSNNSSHH14 pKa = 6.12YY15 pKa = 10.27LAIAVVCSEE24 pKa = 4.35NSDD27 pKa = 3.72EE28 pKa = 5.07LLMFEE33 pKa = 5.68SEE35 pKa = 4.72DD36 pKa = 3.74SNEE39 pKa = 3.74NAIFFEE45 pKa = 4.65DD46 pKa = 3.5SCVISKK52 pKa = 10.57SPSGGALNCAEE63 pKa = 4.25NYY65 pKa = 10.53YY66 pKa = 9.23LTSNAGLLDD75 pKa = 3.84FLKK78 pKa = 9.95STNFKK83 pKa = 10.48SPKK86 pKa = 8.36PLPVANRR93 pKa = 11.84CSNGPNHH100 pKa = 6.45VDD102 pKa = 3.46FHH104 pKa = 6.01MRR106 pKa = 11.84PLHH109 pKa = 6.26LGLIFRR115 pKa = 11.84VNLSLKK121 pKa = 10.76SLDD124 pKa = 5.13DD125 pKa = 4.5LDD127 pKa = 4.28TFQDD131 pKa = 3.61SLVKK135 pKa = 10.56IKK137 pKa = 10.67GFWNNGAQHH146 pKa = 6.17QSMFHH151 pKa = 5.64YY152 pKa = 10.52LSNFINYY159 pKa = 7.17EE160 pKa = 3.81KK161 pKa = 10.99YY162 pKa = 10.48SIVSFEE168 pKa = 3.93YY169 pKa = 10.38FKK171 pKa = 11.45LL172 pKa = 3.79
MM1 pKa = 7.41LSDD4 pKa = 4.21YY5 pKa = 10.96EE6 pKa = 4.37VGSNNSSHH14 pKa = 6.12YY15 pKa = 10.27LAIAVVCSEE24 pKa = 4.35NSDD27 pKa = 3.72EE28 pKa = 5.07LLMFEE33 pKa = 5.68SEE35 pKa = 4.72DD36 pKa = 3.74SNEE39 pKa = 3.74NAIFFEE45 pKa = 4.65DD46 pKa = 3.5SCVISKK52 pKa = 10.57SPSGGALNCAEE63 pKa = 4.25NYY65 pKa = 10.53YY66 pKa = 9.23LTSNAGLLDD75 pKa = 3.84FLKK78 pKa = 9.95STNFKK83 pKa = 10.48SPKK86 pKa = 8.36PLPVANRR93 pKa = 11.84CSNGPNHH100 pKa = 6.45VDD102 pKa = 3.46FHH104 pKa = 6.01MRR106 pKa = 11.84PLHH109 pKa = 6.26LGLIFRR115 pKa = 11.84VNLSLKK121 pKa = 10.76SLDD124 pKa = 5.13DD125 pKa = 4.5LDD127 pKa = 4.28TFQDD131 pKa = 3.61SLVKK135 pKa = 10.56IKK137 pKa = 10.67GFWNNGAQHH146 pKa = 6.17QSMFHH151 pKa = 5.64YY152 pKa = 10.52LSNFINYY159 pKa = 7.17EE160 pKa = 3.81KK161 pKa = 10.99YY162 pKa = 10.48SIVSFEE168 pKa = 3.93YY169 pKa = 10.38FKK171 pKa = 11.45LL172 pKa = 3.79
Molecular weight: 19.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1HSF4|A0A0K1HSF4_9CLOS CP OS=Tobacco virus 1 OX=1692045 GN=CP PE=4 SV=1
MM1 pKa = 7.76MITLISVFSTIRR13 pKa = 11.84SQAIPPRR20 pKa = 11.84SPSLQEE26 pKa = 3.55NLYY29 pKa = 10.6SFEE32 pKa = 3.87SRR34 pKa = 11.84NYY36 pKa = 10.77NFLTCEE42 pKa = 4.29RR43 pKa = 11.84YY44 pKa = 9.77SSPTVFGKK52 pKa = 10.73AMARR56 pKa = 11.84NLIEE60 pKa = 4.59RR61 pKa = 11.84CFTTDD66 pKa = 3.77KK67 pKa = 10.86FLEE70 pKa = 4.71FKK72 pKa = 10.61NSPIRR77 pKa = 11.84FSDD80 pKa = 3.6SALLKK85 pKa = 10.09WMQKK89 pKa = 9.78RR90 pKa = 11.84DD91 pKa = 3.35SSQIKK96 pKa = 9.9ALQSEE101 pKa = 4.79LTRR104 pKa = 11.84PLDD107 pKa = 3.56LSTAVHH113 pKa = 5.84YY114 pKa = 9.28FKK116 pKa = 11.44LMVKK120 pKa = 9.86RR121 pKa = 11.84DD122 pKa = 3.82AKK124 pKa = 11.02VKK126 pKa = 10.62LDD128 pKa = 3.62STCLTKK134 pKa = 10.64HH135 pKa = 5.98SAAQNIMFHH144 pKa = 6.39AKK146 pKa = 9.62AVNALYY152 pKa = 10.49SPCFDD157 pKa = 3.41EE158 pKa = 5.14LKK160 pKa = 10.98NRR162 pKa = 11.84FLSSLKK168 pKa = 9.6KK169 pKa = 9.95HH170 pKa = 5.64IVFFTEE176 pKa = 3.18MDD178 pKa = 3.03NRR180 pKa = 11.84TFARR184 pKa = 11.84VVNALVGSDD193 pKa = 4.85DD194 pKa = 3.85SDD196 pKa = 3.82LHH198 pKa = 7.57VGEE201 pKa = 4.56VDD203 pKa = 4.23FSKK206 pKa = 10.66FDD208 pKa = 3.29KK209 pKa = 11.13SQDD212 pKa = 2.91IFIKK216 pKa = 9.44EE217 pKa = 4.12FEE219 pKa = 4.35RR220 pKa = 11.84EE221 pKa = 4.23VYY223 pKa = 9.62TLLGFDD229 pKa = 4.62EE230 pKa = 5.56EE231 pKa = 4.99MLQLWMEE238 pKa = 4.4GEE240 pKa = 4.43YY241 pKa = 10.33SAKK244 pKa = 9.9ATTLDD249 pKa = 3.4GKK251 pKa = 11.2LSFEE255 pKa = 4.61VKK257 pKa = 8.97NQRR260 pKa = 11.84RR261 pKa = 11.84SGASNTWIGNSIVTLGILSMYY282 pKa = 10.23YY283 pKa = 10.31KK284 pKa = 10.43VDD286 pKa = 3.25EE287 pKa = 4.48LLALFVSGDD296 pKa = 3.63DD297 pKa = 3.5SLMYY301 pKa = 10.52SNKK304 pKa = 9.94PIANYY309 pKa = 10.47AEE311 pKa = 4.65SICVEE316 pKa = 4.2TGFEE320 pKa = 4.39TKK322 pKa = 10.0FLSPSVPYY330 pKa = 9.82FCSKK334 pKa = 10.44FVVHH338 pKa = 6.82CGFKK342 pKa = 10.3TFFVPDD348 pKa = 4.28PYY350 pKa = 11.75KK351 pKa = 11.22LMVKK355 pKa = 10.18LGSVRR360 pKa = 11.84KK361 pKa = 9.31EE362 pKa = 3.87LTDD365 pKa = 3.45KK366 pKa = 11.32DD367 pKa = 3.84LFEE370 pKa = 4.84VFTSFKK376 pKa = 11.11DD377 pKa = 3.51LTKK380 pKa = 10.96DD381 pKa = 3.27LGDD384 pKa = 3.49EE385 pKa = 4.13RR386 pKa = 11.84VLEE389 pKa = 4.03KK390 pKa = 11.24LNLLMIAKK398 pKa = 9.81YY399 pKa = 10.46GYY401 pKa = 10.54NSDD404 pKa = 3.66FALPALRR411 pKa = 11.84SIHH414 pKa = 6.36CLASNFSSFSKK425 pKa = 10.57LFEE428 pKa = 4.79KK429 pKa = 9.75STGWVVVPKK438 pKa = 10.72LNSYY442 pKa = 8.38YY443 pKa = 10.72KK444 pKa = 10.65KK445 pKa = 10.75LIALGAYY452 pKa = 8.16NEE454 pKa = 4.57RR455 pKa = 11.84YY456 pKa = 7.08VTPFGEE462 pKa = 4.57QYY464 pKa = 10.11FVAWW468 pKa = 3.73
MM1 pKa = 7.76MITLISVFSTIRR13 pKa = 11.84SQAIPPRR20 pKa = 11.84SPSLQEE26 pKa = 3.55NLYY29 pKa = 10.6SFEE32 pKa = 3.87SRR34 pKa = 11.84NYY36 pKa = 10.77NFLTCEE42 pKa = 4.29RR43 pKa = 11.84YY44 pKa = 9.77SSPTVFGKK52 pKa = 10.73AMARR56 pKa = 11.84NLIEE60 pKa = 4.59RR61 pKa = 11.84CFTTDD66 pKa = 3.77KK67 pKa = 10.86FLEE70 pKa = 4.71FKK72 pKa = 10.61NSPIRR77 pKa = 11.84FSDD80 pKa = 3.6SALLKK85 pKa = 10.09WMQKK89 pKa = 9.78RR90 pKa = 11.84DD91 pKa = 3.35SSQIKK96 pKa = 9.9ALQSEE101 pKa = 4.79LTRR104 pKa = 11.84PLDD107 pKa = 3.56LSTAVHH113 pKa = 5.84YY114 pKa = 9.28FKK116 pKa = 11.44LMVKK120 pKa = 9.86RR121 pKa = 11.84DD122 pKa = 3.82AKK124 pKa = 11.02VKK126 pKa = 10.62LDD128 pKa = 3.62STCLTKK134 pKa = 10.64HH135 pKa = 5.98SAAQNIMFHH144 pKa = 6.39AKK146 pKa = 9.62AVNALYY152 pKa = 10.49SPCFDD157 pKa = 3.41EE158 pKa = 5.14LKK160 pKa = 10.98NRR162 pKa = 11.84FLSSLKK168 pKa = 9.6KK169 pKa = 9.95HH170 pKa = 5.64IVFFTEE176 pKa = 3.18MDD178 pKa = 3.03NRR180 pKa = 11.84TFARR184 pKa = 11.84VVNALVGSDD193 pKa = 4.85DD194 pKa = 3.85SDD196 pKa = 3.82LHH198 pKa = 7.57VGEE201 pKa = 4.56VDD203 pKa = 4.23FSKK206 pKa = 10.66FDD208 pKa = 3.29KK209 pKa = 11.13SQDD212 pKa = 2.91IFIKK216 pKa = 9.44EE217 pKa = 4.12FEE219 pKa = 4.35RR220 pKa = 11.84EE221 pKa = 4.23VYY223 pKa = 9.62TLLGFDD229 pKa = 4.62EE230 pKa = 5.56EE231 pKa = 4.99MLQLWMEE238 pKa = 4.4GEE240 pKa = 4.43YY241 pKa = 10.33SAKK244 pKa = 9.9ATTLDD249 pKa = 3.4GKK251 pKa = 11.2LSFEE255 pKa = 4.61VKK257 pKa = 8.97NQRR260 pKa = 11.84RR261 pKa = 11.84SGASNTWIGNSIVTLGILSMYY282 pKa = 10.23YY283 pKa = 10.31KK284 pKa = 10.43VDD286 pKa = 3.25EE287 pKa = 4.48LLALFVSGDD296 pKa = 3.63DD297 pKa = 3.5SLMYY301 pKa = 10.52SNKK304 pKa = 9.94PIANYY309 pKa = 10.47AEE311 pKa = 4.65SICVEE316 pKa = 4.2TGFEE320 pKa = 4.39TKK322 pKa = 10.0FLSPSVPYY330 pKa = 9.82FCSKK334 pKa = 10.44FVVHH338 pKa = 6.82CGFKK342 pKa = 10.3TFFVPDD348 pKa = 4.28PYY350 pKa = 11.75KK351 pKa = 11.22LMVKK355 pKa = 10.18LGSVRR360 pKa = 11.84KK361 pKa = 9.31EE362 pKa = 3.87LTDD365 pKa = 3.45KK366 pKa = 11.32DD367 pKa = 3.84LFEE370 pKa = 4.84VFTSFKK376 pKa = 11.11DD377 pKa = 3.51LTKK380 pKa = 10.96DD381 pKa = 3.27LGDD384 pKa = 3.49EE385 pKa = 4.13RR386 pKa = 11.84VLEE389 pKa = 4.03KK390 pKa = 11.24LNLLMIAKK398 pKa = 9.81YY399 pKa = 10.46GYY401 pKa = 10.54NSDD404 pKa = 3.66FALPALRR411 pKa = 11.84SIHH414 pKa = 6.36CLASNFSSFSKK425 pKa = 10.57LFEE428 pKa = 4.79KK429 pKa = 9.75STGWVVVPKK438 pKa = 10.72LNSYY442 pKa = 8.38YY443 pKa = 10.72KK444 pKa = 10.65KK445 pKa = 10.75LIALGAYY452 pKa = 8.16NEE454 pKa = 4.57RR455 pKa = 11.84YY456 pKa = 7.08VTPFGEE462 pKa = 4.57QYY464 pKa = 10.11FVAWW468 pKa = 3.73
Molecular weight: 53.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4936 |
65 |
2471 |
548.4 |
62.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.592 ± 0.267 | 2.431 ± 0.343 |
5.794 ± 0.156 | 6.22 ± 0.272 |
5.693 ± 0.356 | 4.862 ± 0.435 |
2.168 ± 0.238 | 5.369 ± 0.287 |
6.382 ± 0.328 | 9.542 ± 0.526 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.026 ± 0.133 | 4.781 ± 0.417 |
3.383 ± 0.195 | 2.208 ± 0.143 |
5.916 ± 0.59 | 10.008 ± 0.521 |
5.429 ± 0.367 | 7.78 ± 0.605 |
0.527 ± 0.141 | 3.89 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
