
Gracilimonas sp. 8A47
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Rhodohalobacter
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
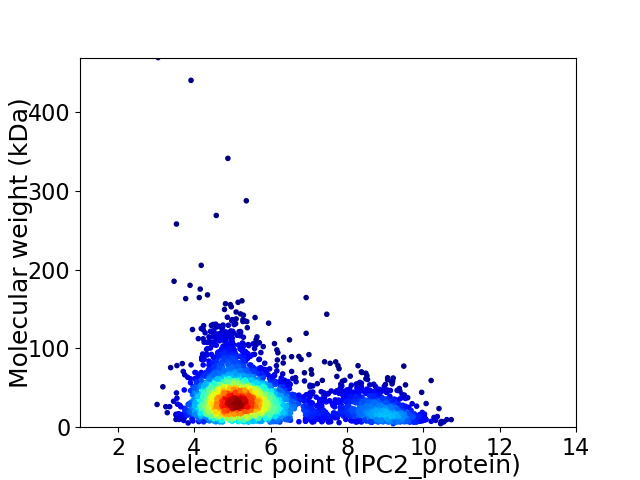
Virtual 2D-PAGE plot for 3239 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
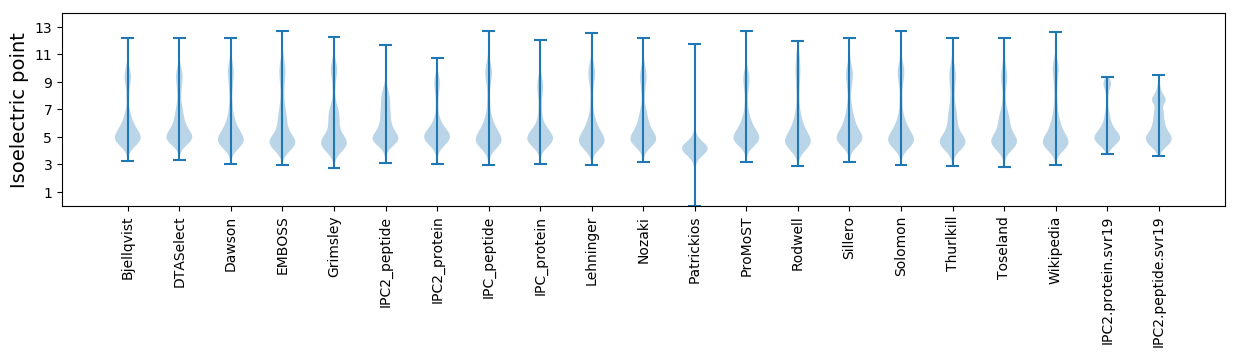
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316TZG5|A0A316TZG5_9BACT Arginase OS=Gracilimonas sp. 8A47 OX=2079485 GN=DDZ15_12985 PE=3 SV=1
MM1 pKa = 7.34KK2 pKa = 9.96RR3 pKa = 11.84YY4 pKa = 10.29SILTLLIFSTAVLMHH19 pKa = 6.81GCTDD23 pKa = 4.62DD24 pKa = 5.16GPLGPDD30 pKa = 3.46PDD32 pKa = 4.59DD33 pKa = 4.53FDD35 pKa = 5.66RR36 pKa = 11.84SAILTNWADD45 pKa = 3.67NLIVPAYY52 pKa = 9.93QNFVADD58 pKa = 4.57AEE60 pKa = 4.38ALEE63 pKa = 4.4SAAADD68 pKa = 4.83FSANPAADD76 pKa = 3.6EE77 pKa = 4.27LLALRR82 pKa = 11.84NAFEE86 pKa = 3.9TAYY89 pKa = 11.11LSFQKK94 pKa = 10.81VSMYY98 pKa = 10.52EE99 pKa = 3.64IGPAMQVGAQGINLRR114 pKa = 11.84NYY116 pKa = 9.66LNSYY120 pKa = 10.01PSDD123 pKa = 3.21TAQIVEE129 pKa = 4.36NVSLNDD135 pKa = 3.42INLDD139 pKa = 4.03LPSQLDD145 pKa = 3.69AQGFPALDD153 pKa = 3.49YY154 pKa = 11.07LLYY157 pKa = 10.69GAAGSEE163 pKa = 4.19AEE165 pKa = 4.13ILALYY170 pKa = 10.51SEE172 pKa = 5.16GADD175 pKa = 3.16APLYY179 pKa = 10.32RR180 pKa = 11.84SYY182 pKa = 11.31LQTLSARR189 pKa = 11.84IAGLAGNVLNAWEE202 pKa = 4.07TGYY205 pKa = 10.88RR206 pKa = 11.84DD207 pKa = 3.67EE208 pKa = 4.98FTDD211 pKa = 4.1NSGNGANASIDD222 pKa = 3.57MMVNDD227 pKa = 4.22YY228 pKa = 10.97IFYY231 pKa = 8.77YY232 pKa = 9.89EE233 pKa = 3.5KK234 pKa = 9.89WLRR237 pKa = 11.84AGKK240 pKa = 10.23VGIPAGVFSGTPLPTHH256 pKa = 6.83VEE258 pKa = 3.9ALYY261 pKa = 10.48HH262 pKa = 6.33GSFSRR267 pKa = 11.84QLALEE272 pKa = 4.32ALSAVQDD279 pKa = 4.04FFNGNHH285 pKa = 5.28VNGTGSGEE293 pKa = 4.28SLNSYY298 pKa = 10.75LDD300 pKa = 3.78YY301 pKa = 11.08LDD303 pKa = 3.79SRR305 pKa = 11.84RR306 pKa = 11.84DD307 pKa = 3.55GSQLSVLINSQFEE320 pKa = 4.32SARR323 pKa = 11.84SEE325 pKa = 4.7LEE327 pKa = 3.84SLQQNFADD335 pKa = 4.05QVEE338 pKa = 4.24NDD340 pKa = 4.06NIRR343 pKa = 11.84MLNAYY348 pKa = 9.96DD349 pKa = 4.03EE350 pKa = 4.38LQKK353 pKa = 11.17NVVFMKK359 pKa = 10.61VDD361 pKa = 3.29MLQALNISVDD371 pKa = 3.76YY372 pKa = 11.37VDD374 pKa = 5.38ADD376 pKa = 3.45GDD378 pKa = 3.83
MM1 pKa = 7.34KK2 pKa = 9.96RR3 pKa = 11.84YY4 pKa = 10.29SILTLLIFSTAVLMHH19 pKa = 6.81GCTDD23 pKa = 4.62DD24 pKa = 5.16GPLGPDD30 pKa = 3.46PDD32 pKa = 4.59DD33 pKa = 4.53FDD35 pKa = 5.66RR36 pKa = 11.84SAILTNWADD45 pKa = 3.67NLIVPAYY52 pKa = 9.93QNFVADD58 pKa = 4.57AEE60 pKa = 4.38ALEE63 pKa = 4.4SAAADD68 pKa = 4.83FSANPAADD76 pKa = 3.6EE77 pKa = 4.27LLALRR82 pKa = 11.84NAFEE86 pKa = 3.9TAYY89 pKa = 11.11LSFQKK94 pKa = 10.81VSMYY98 pKa = 10.52EE99 pKa = 3.64IGPAMQVGAQGINLRR114 pKa = 11.84NYY116 pKa = 9.66LNSYY120 pKa = 10.01PSDD123 pKa = 3.21TAQIVEE129 pKa = 4.36NVSLNDD135 pKa = 3.42INLDD139 pKa = 4.03LPSQLDD145 pKa = 3.69AQGFPALDD153 pKa = 3.49YY154 pKa = 11.07LLYY157 pKa = 10.69GAAGSEE163 pKa = 4.19AEE165 pKa = 4.13ILALYY170 pKa = 10.51SEE172 pKa = 5.16GADD175 pKa = 3.16APLYY179 pKa = 10.32RR180 pKa = 11.84SYY182 pKa = 11.31LQTLSARR189 pKa = 11.84IAGLAGNVLNAWEE202 pKa = 4.07TGYY205 pKa = 10.88RR206 pKa = 11.84DD207 pKa = 3.67EE208 pKa = 4.98FTDD211 pKa = 4.1NSGNGANASIDD222 pKa = 3.57MMVNDD227 pKa = 4.22YY228 pKa = 10.97IFYY231 pKa = 8.77YY232 pKa = 9.89EE233 pKa = 3.5KK234 pKa = 9.89WLRR237 pKa = 11.84AGKK240 pKa = 10.23VGIPAGVFSGTPLPTHH256 pKa = 6.83VEE258 pKa = 3.9ALYY261 pKa = 10.48HH262 pKa = 6.33GSFSRR267 pKa = 11.84QLALEE272 pKa = 4.32ALSAVQDD279 pKa = 4.04FFNGNHH285 pKa = 5.28VNGTGSGEE293 pKa = 4.28SLNSYY298 pKa = 10.75LDD300 pKa = 3.78YY301 pKa = 11.08LDD303 pKa = 3.79SRR305 pKa = 11.84RR306 pKa = 11.84DD307 pKa = 3.55GSQLSVLINSQFEE320 pKa = 4.32SARR323 pKa = 11.84SEE325 pKa = 4.7LEE327 pKa = 3.84SLQQNFADD335 pKa = 4.05QVEE338 pKa = 4.24NDD340 pKa = 4.06NIRR343 pKa = 11.84MLNAYY348 pKa = 9.96DD349 pKa = 4.03EE350 pKa = 4.38LQKK353 pKa = 11.17NVVFMKK359 pKa = 10.61VDD361 pKa = 3.29MLQALNISVDD371 pKa = 3.76YY372 pKa = 11.37VDD374 pKa = 5.38ADD376 pKa = 3.45GDD378 pKa = 3.83
Molecular weight: 41.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316TVQ0|A0A316TVQ0_9BACT Uncharacterized protein OS=Gracilimonas sp. 8A47 OX=2079485 GN=DDZ15_08730 PE=4 SV=1
MM1 pKa = 7.34WKK3 pKa = 9.8SVPRR7 pKa = 11.84FRR9 pKa = 11.84QSILVLGGLFLTLIWLIILSVGIGAVPISPSQVISIFLNQAGIEE53 pKa = 4.09TGLGYY58 pKa = 10.78EE59 pKa = 4.54SVQEE63 pKa = 4.26SILLNIRR70 pKa = 11.84LPRR73 pKa = 11.84VLLAVLVGAALAISGAAMQGLFRR96 pKa = 11.84NPLADD101 pKa = 4.27PGLIGVSSGAALATAIAMVVLSSLAWPFLDD131 pKa = 5.03LLGEE135 pKa = 4.1ALIPLAAFAGGTSATILVYY154 pKa = 10.49RR155 pKa = 11.84LSTSRR160 pKa = 11.84GRR162 pKa = 11.84TNVATMLLAGIAINAMAGALIGFMIFLADD191 pKa = 4.26DD192 pKa = 3.95DD193 pKa = 4.13QLRR196 pKa = 11.84DD197 pKa = 3.57LTFWTLGSLGGAMWKK212 pKa = 10.08SVWIVLPILMAAILFLPRR230 pKa = 11.84LSKK233 pKa = 10.75GLNAILLGEE242 pKa = 4.25AEE244 pKa = 4.1ARR246 pKa = 11.84HH247 pKa = 5.85LGVRR251 pKa = 11.84VQRR254 pKa = 11.84LKK256 pKa = 10.78KK257 pKa = 10.47VIIIFVGLAVGAAVSVSGMIGFVGLVVPHH286 pKa = 7.34ILRR289 pKa = 11.84LWIGPDD295 pKa = 3.23HH296 pKa = 7.05RR297 pKa = 11.84FLMPGSAILGGLLLLASDD315 pKa = 4.43LAARR319 pKa = 11.84TMVAPAEE326 pKa = 4.07LPIGVITASIGAPFFLWLLLRR347 pKa = 11.84NRR349 pKa = 11.84EE350 pKa = 3.98LRR352 pKa = 11.84GYY354 pKa = 10.24LL355 pKa = 3.48
MM1 pKa = 7.34WKK3 pKa = 9.8SVPRR7 pKa = 11.84FRR9 pKa = 11.84QSILVLGGLFLTLIWLIILSVGIGAVPISPSQVISIFLNQAGIEE53 pKa = 4.09TGLGYY58 pKa = 10.78EE59 pKa = 4.54SVQEE63 pKa = 4.26SILLNIRR70 pKa = 11.84LPRR73 pKa = 11.84VLLAVLVGAALAISGAAMQGLFRR96 pKa = 11.84NPLADD101 pKa = 4.27PGLIGVSSGAALATAIAMVVLSSLAWPFLDD131 pKa = 5.03LLGEE135 pKa = 4.1ALIPLAAFAGGTSATILVYY154 pKa = 10.49RR155 pKa = 11.84LSTSRR160 pKa = 11.84GRR162 pKa = 11.84TNVATMLLAGIAINAMAGALIGFMIFLADD191 pKa = 4.26DD192 pKa = 3.95DD193 pKa = 4.13QLRR196 pKa = 11.84DD197 pKa = 3.57LTFWTLGSLGGAMWKK212 pKa = 10.08SVWIVLPILMAAILFLPRR230 pKa = 11.84LSKK233 pKa = 10.75GLNAILLGEE242 pKa = 4.25AEE244 pKa = 4.1ARR246 pKa = 11.84HH247 pKa = 5.85LGVRR251 pKa = 11.84VQRR254 pKa = 11.84LKK256 pKa = 10.78KK257 pKa = 10.47VIIIFVGLAVGAAVSVSGMIGFVGLVVPHH286 pKa = 7.34ILRR289 pKa = 11.84LWIGPDD295 pKa = 3.23HH296 pKa = 7.05RR297 pKa = 11.84FLMPGSAILGGLLLLASDD315 pKa = 4.43LAARR319 pKa = 11.84TMVAPAEE326 pKa = 4.07LPIGVITASIGAPFFLWLLLRR347 pKa = 11.84NRR349 pKa = 11.84EE350 pKa = 3.98LRR352 pKa = 11.84GYY354 pKa = 10.24LL355 pKa = 3.48
Molecular weight: 37.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1132565 |
38 |
4770 |
349.7 |
39.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.192 ± 0.05 | 0.607 ± 0.012 |
6.016 ± 0.034 | 7.393 ± 0.051 |
4.695 ± 0.031 | 7.283 ± 0.039 |
1.965 ± 0.023 | 6.845 ± 0.041 |
4.74 ± 0.052 | 9.522 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.55 ± 0.023 | 4.528 ± 0.035 |
4.142 ± 0.023 | 3.424 ± 0.025 |
5.4 ± 0.038 | 6.967 ± 0.039 |
5.521 ± 0.05 | 6.482 ± 0.035 |
1.232 ± 0.016 | 3.496 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
