
Human feces pecovirus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
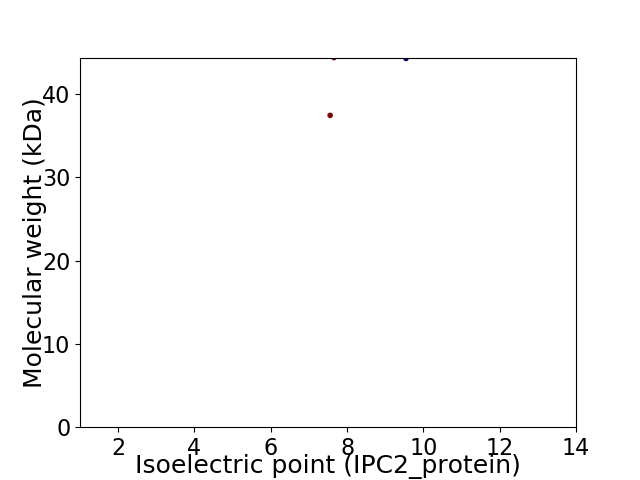
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142J7H6|A0A142J7H6_9VIRU Replication associated protein OS=Human feces pecovirus OX=1820160 PE=4 SV=1
MM1 pKa = 8.1SMRR4 pKa = 11.84TRR6 pKa = 11.84NWCVTSWTCPDD17 pKa = 3.48YY18 pKa = 11.54DD19 pKa = 3.69SVEE22 pKa = 4.1RR23 pKa = 11.84LVVSGEE29 pKa = 3.88YY30 pKa = 10.34KK31 pKa = 9.82YY32 pKa = 11.36VCVGKK37 pKa = 7.87EE38 pKa = 3.72TCPRR42 pKa = 11.84TGKK45 pKa = 8.4VHH47 pKa = 4.87WHH49 pKa = 6.83WYY51 pKa = 7.75LQAVNKK57 pKa = 10.5KK58 pKa = 8.55SMRR61 pKa = 11.84QIKK64 pKa = 10.17KK65 pKa = 10.46DD66 pKa = 3.21VGDD69 pKa = 4.03RR70 pKa = 11.84MAHH73 pKa = 6.13AEE75 pKa = 4.09AANGPADD82 pKa = 3.98AGIAYY87 pKa = 9.08VKK89 pKa = 10.61KK90 pKa = 10.5DD91 pKa = 3.85GEE93 pKa = 4.18WKK95 pKa = 10.18EE96 pKa = 3.61WGEE99 pKa = 3.88PKK101 pKa = 10.26KK102 pKa = 10.53QGRR105 pKa = 11.84RR106 pKa = 11.84SDD108 pKa = 5.02LEE110 pKa = 4.14DD111 pKa = 3.49CKK113 pKa = 11.07KK114 pKa = 10.79IIDD117 pKa = 4.05NSVDD121 pKa = 3.46DD122 pKa = 5.64AMGTMQQIADD132 pKa = 3.6THH134 pKa = 6.26FGDD137 pKa = 4.54FVRR140 pKa = 11.84YY141 pKa = 8.38YY142 pKa = 10.81RR143 pKa = 11.84GFAKK147 pKa = 10.45YY148 pKa = 11.04ADD150 pKa = 3.54MVDD153 pKa = 3.2IKK155 pKa = 10.43RR156 pKa = 11.84AKK158 pKa = 9.3QRR160 pKa = 11.84EE161 pKa = 3.95PHH163 pKa = 5.31MPEE166 pKa = 3.66VIVYY170 pKa = 9.46LGRR173 pKa = 11.84AGSGKK178 pKa = 10.23SYY180 pKa = 10.44HH181 pKa = 6.94CYY183 pKa = 9.89HH184 pKa = 7.49DD185 pKa = 3.73EE186 pKa = 4.63GYY188 pKa = 10.22QRR190 pKa = 11.84SGYY193 pKa = 9.76RR194 pKa = 11.84YY195 pKa = 8.83LVQMEE200 pKa = 4.56SKK202 pKa = 9.53TYY204 pKa = 9.65FDD206 pKa = 5.07GYY208 pKa = 10.85DD209 pKa = 3.49GEE211 pKa = 4.34KK212 pKa = 10.21TIWFDD217 pKa = 3.52EE218 pKa = 4.47FSGRR222 pKa = 11.84VMSFTVFTQIVDD234 pKa = 3.28QWGSRR239 pKa = 11.84VEE241 pKa = 4.24TKK243 pKa = 10.55GGSRR247 pKa = 11.84QIFPEE252 pKa = 4.1RR253 pKa = 11.84FLISTVEE260 pKa = 4.2WPSQWWAGSSRR271 pKa = 11.84FNADD275 pKa = 2.9PYY277 pKa = 10.24QLYY280 pKa = 10.41RR281 pKa = 11.84RR282 pKa = 11.84ITKK285 pKa = 9.81IYY287 pKa = 7.71WCRR290 pKa = 11.84GPGQEE295 pKa = 4.43PVEE298 pKa = 4.14LKK300 pKa = 10.62KK301 pKa = 10.96DD302 pKa = 2.85GWTCKK307 pKa = 10.78NIDD310 pKa = 3.73EE311 pKa = 4.34YY312 pKa = 11.62EE313 pKa = 4.1EE314 pKa = 5.01LFGKK318 pKa = 10.62LPP320 pKa = 3.67
MM1 pKa = 8.1SMRR4 pKa = 11.84TRR6 pKa = 11.84NWCVTSWTCPDD17 pKa = 3.48YY18 pKa = 11.54DD19 pKa = 3.69SVEE22 pKa = 4.1RR23 pKa = 11.84LVVSGEE29 pKa = 3.88YY30 pKa = 10.34KK31 pKa = 9.82YY32 pKa = 11.36VCVGKK37 pKa = 7.87EE38 pKa = 3.72TCPRR42 pKa = 11.84TGKK45 pKa = 8.4VHH47 pKa = 4.87WHH49 pKa = 6.83WYY51 pKa = 7.75LQAVNKK57 pKa = 10.5KK58 pKa = 8.55SMRR61 pKa = 11.84QIKK64 pKa = 10.17KK65 pKa = 10.46DD66 pKa = 3.21VGDD69 pKa = 4.03RR70 pKa = 11.84MAHH73 pKa = 6.13AEE75 pKa = 4.09AANGPADD82 pKa = 3.98AGIAYY87 pKa = 9.08VKK89 pKa = 10.61KK90 pKa = 10.5DD91 pKa = 3.85GEE93 pKa = 4.18WKK95 pKa = 10.18EE96 pKa = 3.61WGEE99 pKa = 3.88PKK101 pKa = 10.26KK102 pKa = 10.53QGRR105 pKa = 11.84RR106 pKa = 11.84SDD108 pKa = 5.02LEE110 pKa = 4.14DD111 pKa = 3.49CKK113 pKa = 11.07KK114 pKa = 10.79IIDD117 pKa = 4.05NSVDD121 pKa = 3.46DD122 pKa = 5.64AMGTMQQIADD132 pKa = 3.6THH134 pKa = 6.26FGDD137 pKa = 4.54FVRR140 pKa = 11.84YY141 pKa = 8.38YY142 pKa = 10.81RR143 pKa = 11.84GFAKK147 pKa = 10.45YY148 pKa = 11.04ADD150 pKa = 3.54MVDD153 pKa = 3.2IKK155 pKa = 10.43RR156 pKa = 11.84AKK158 pKa = 9.3QRR160 pKa = 11.84EE161 pKa = 3.95PHH163 pKa = 5.31MPEE166 pKa = 3.66VIVYY170 pKa = 9.46LGRR173 pKa = 11.84AGSGKK178 pKa = 10.23SYY180 pKa = 10.44HH181 pKa = 6.94CYY183 pKa = 9.89HH184 pKa = 7.49DD185 pKa = 3.73EE186 pKa = 4.63GYY188 pKa = 10.22QRR190 pKa = 11.84SGYY193 pKa = 9.76RR194 pKa = 11.84YY195 pKa = 8.83LVQMEE200 pKa = 4.56SKK202 pKa = 9.53TYY204 pKa = 9.65FDD206 pKa = 5.07GYY208 pKa = 10.85DD209 pKa = 3.49GEE211 pKa = 4.34KK212 pKa = 10.21TIWFDD217 pKa = 3.52EE218 pKa = 4.47FSGRR222 pKa = 11.84VMSFTVFTQIVDD234 pKa = 3.28QWGSRR239 pKa = 11.84VEE241 pKa = 4.24TKK243 pKa = 10.55GGSRR247 pKa = 11.84QIFPEE252 pKa = 4.1RR253 pKa = 11.84FLISTVEE260 pKa = 4.2WPSQWWAGSSRR271 pKa = 11.84FNADD275 pKa = 2.9PYY277 pKa = 10.24QLYY280 pKa = 10.41RR281 pKa = 11.84RR282 pKa = 11.84ITKK285 pKa = 9.81IYY287 pKa = 7.71WCRR290 pKa = 11.84GPGQEE295 pKa = 4.43PVEE298 pKa = 4.14LKK300 pKa = 10.62KK301 pKa = 10.96DD302 pKa = 2.85GWTCKK307 pKa = 10.78NIDD310 pKa = 3.73EE311 pKa = 4.34YY312 pKa = 11.62EE313 pKa = 4.1EE314 pKa = 5.01LFGKK318 pKa = 10.62LPP320 pKa = 3.67
Molecular weight: 37.42 kDa
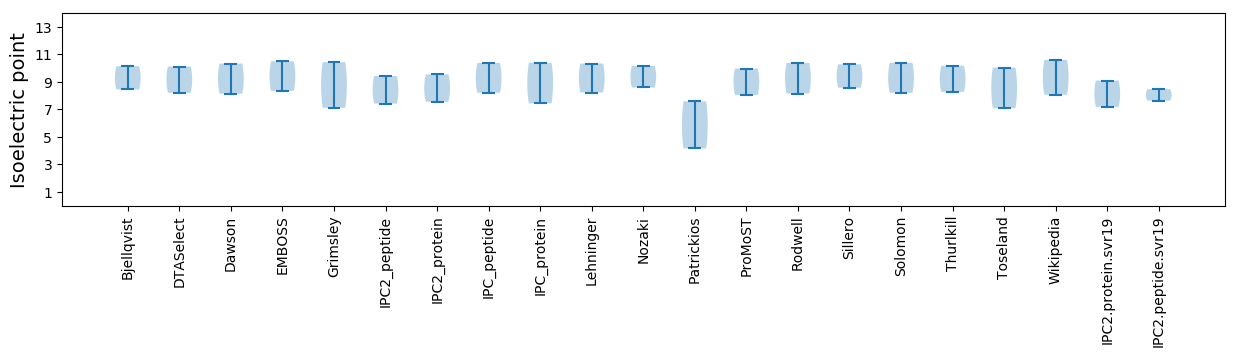
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142J7H6|A0A142J7H6_9VIRU Replication associated protein OS=Human feces pecovirus OX=1820160 PE=4 SV=1
MM1 pKa = 7.12SHH3 pKa = 6.93LVSHH7 pKa = 6.82INVMARR13 pKa = 11.84RR14 pKa = 11.84YY15 pKa = 8.75RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FGRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.0LRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ALLRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LLRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGRR51 pKa = 11.84RR52 pKa = 11.84FTVSVQRR59 pKa = 11.84DD60 pKa = 3.58FALIAASRR68 pKa = 11.84VPEE71 pKa = 4.05ATQLAGNGTNVAGPGGVVGIDD92 pKa = 2.95TGGFRR97 pKa = 11.84SYY99 pKa = 11.36SVLALPPDD107 pKa = 3.95WIEE110 pKa = 3.82RR111 pKa = 11.84TVGDD115 pKa = 3.64TRR117 pKa = 11.84LVGNQWDD124 pKa = 3.75VSNKK128 pKa = 9.57YY129 pKa = 9.05LQRR132 pKa = 11.84CIGVGTGEE140 pKa = 4.27SGMLTDD146 pKa = 5.27LPQIIWSKK154 pKa = 9.66PLPVNFNCGSLWDD167 pKa = 3.74LCEE170 pKa = 4.08EE171 pKa = 4.54YY172 pKa = 10.5RR173 pKa = 11.84IAWIALSFTVAEE185 pKa = 4.42SQTKK189 pKa = 9.42NNRR192 pKa = 11.84HH193 pKa = 6.43LYY195 pKa = 10.49LEE197 pKa = 4.33WTNLPSAKK205 pKa = 10.19APQWTDD211 pKa = 2.63LDD213 pKa = 4.29GMVLPEE219 pKa = 4.98GYY221 pKa = 10.5AGGGTNVDD229 pKa = 3.4TNGFNWLCRR238 pKa = 11.84PIDD241 pKa = 3.47IALACSPSGRR251 pKa = 11.84QSKK254 pKa = 10.44LNGWHH259 pKa = 7.06RR260 pKa = 11.84SILSYY265 pKa = 7.76THH267 pKa = 7.09PVTIRR272 pKa = 11.84YY273 pKa = 8.46RR274 pKa = 11.84PRR276 pKa = 11.84HH277 pKa = 5.85ADD279 pKa = 3.08MTIDD283 pKa = 3.83NNPDD287 pKa = 3.19VAPDD291 pKa = 3.52SSNQNKK297 pKa = 9.65PKK299 pKa = 10.04MIYY302 pKa = 9.88SDD304 pKa = 3.61NFSEE308 pKa = 4.62SGRR311 pKa = 11.84LTRR314 pKa = 11.84GYY316 pKa = 10.74LPVAQASVLLTEE328 pKa = 4.42QCWFGPCIRR337 pKa = 11.84VVDD340 pKa = 4.07ADD342 pKa = 3.9KK343 pKa = 10.91TPQLGTNEE351 pKa = 3.76QAFNSFYY358 pKa = 10.86DD359 pKa = 3.73EE360 pKa = 4.1YY361 pKa = 11.35GIRR364 pKa = 11.84CSMTAVLRR372 pKa = 11.84MRR374 pKa = 11.84AKK376 pKa = 9.26TCNDD380 pKa = 3.14PVFPYY385 pKa = 9.41YY386 pKa = 9.79TVHH389 pKa = 7.07
MM1 pKa = 7.12SHH3 pKa = 6.93LVSHH7 pKa = 6.82INVMARR13 pKa = 11.84RR14 pKa = 11.84YY15 pKa = 8.75RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FGRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.0LRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ALLRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LLRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGRR51 pKa = 11.84RR52 pKa = 11.84FTVSVQRR59 pKa = 11.84DD60 pKa = 3.58FALIAASRR68 pKa = 11.84VPEE71 pKa = 4.05ATQLAGNGTNVAGPGGVVGIDD92 pKa = 2.95TGGFRR97 pKa = 11.84SYY99 pKa = 11.36SVLALPPDD107 pKa = 3.95WIEE110 pKa = 3.82RR111 pKa = 11.84TVGDD115 pKa = 3.64TRR117 pKa = 11.84LVGNQWDD124 pKa = 3.75VSNKK128 pKa = 9.57YY129 pKa = 9.05LQRR132 pKa = 11.84CIGVGTGEE140 pKa = 4.27SGMLTDD146 pKa = 5.27LPQIIWSKK154 pKa = 9.66PLPVNFNCGSLWDD167 pKa = 3.74LCEE170 pKa = 4.08EE171 pKa = 4.54YY172 pKa = 10.5RR173 pKa = 11.84IAWIALSFTVAEE185 pKa = 4.42SQTKK189 pKa = 9.42NNRR192 pKa = 11.84HH193 pKa = 6.43LYY195 pKa = 10.49LEE197 pKa = 4.33WTNLPSAKK205 pKa = 10.19APQWTDD211 pKa = 2.63LDD213 pKa = 4.29GMVLPEE219 pKa = 4.98GYY221 pKa = 10.5AGGGTNVDD229 pKa = 3.4TNGFNWLCRR238 pKa = 11.84PIDD241 pKa = 3.47IALACSPSGRR251 pKa = 11.84QSKK254 pKa = 10.44LNGWHH259 pKa = 7.06RR260 pKa = 11.84SILSYY265 pKa = 7.76THH267 pKa = 7.09PVTIRR272 pKa = 11.84YY273 pKa = 8.46RR274 pKa = 11.84PRR276 pKa = 11.84HH277 pKa = 5.85ADD279 pKa = 3.08MTIDD283 pKa = 3.83NNPDD287 pKa = 3.19VAPDD291 pKa = 3.52SSNQNKK297 pKa = 9.65PKK299 pKa = 10.04MIYY302 pKa = 9.88SDD304 pKa = 3.61NFSEE308 pKa = 4.62SGRR311 pKa = 11.84LTRR314 pKa = 11.84GYY316 pKa = 10.74LPVAQASVLLTEE328 pKa = 4.42QCWFGPCIRR337 pKa = 11.84VVDD340 pKa = 4.07ADD342 pKa = 3.9KK343 pKa = 10.91TPQLGTNEE351 pKa = 3.76QAFNSFYY358 pKa = 10.86DD359 pKa = 3.73EE360 pKa = 4.1YY361 pKa = 11.35GIRR364 pKa = 11.84CSMTAVLRR372 pKa = 11.84MRR374 pKa = 11.84AKK376 pKa = 9.26TCNDD380 pKa = 3.14PVFPYY385 pKa = 9.41YY386 pKa = 9.79TVHH389 pKa = 7.07
Molecular weight: 44.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
709 |
320 |
389 |
354.5 |
40.84 |
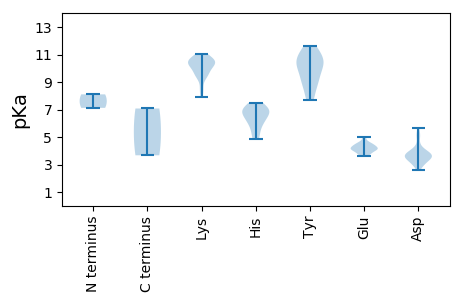
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.783 ± 0.516 | 2.398 ± 0.067 |
6.065 ± 0.739 | 4.795 ± 1.37 |
3.385 ± 0.24 | 8.322 ± 0.282 |
1.975 ± 0.14 | 4.372 ± 0.002 |
4.937 ± 2.1 | 6.065 ± 1.936 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.386 | 3.949 ± 1.366 |
4.937 ± 0.782 | 3.808 ± 0.373 |
9.873 ± 1.769 | 6.347 ± 0.27 |
5.501 ± 0.536 | 6.77 ± 0.069 |
3.244 ± 0.539 | 4.937 ± 0.865 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
