
Robiginitalea myxolifaciens
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Robiginitalea
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
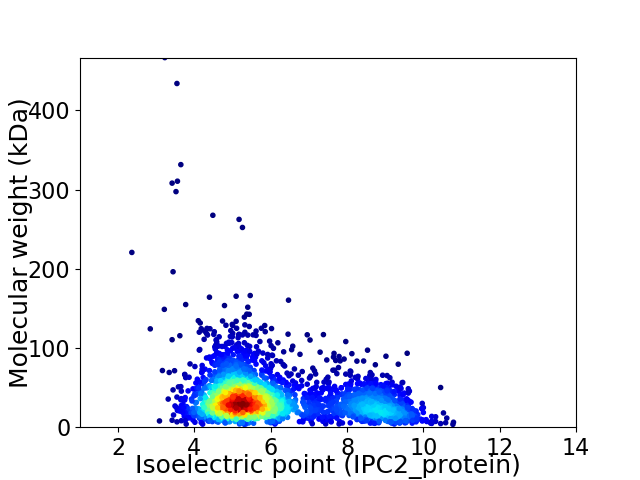
Virtual 2D-PAGE plot for 2844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6HER6|A0A1I6HER6_9FLAO Polyphosphate glucokinase OS=Robiginitalea myxolifaciens OX=400055 GN=SAMN04490243_2660 PE=4 SV=1
MM1 pKa = 6.99TRR3 pKa = 11.84NLVFLLAFLWATYY16 pKa = 10.57SLQAQNSPDD25 pKa = 3.77CRR27 pKa = 11.84SAIPVCADD35 pKa = 3.2APILNFADD43 pKa = 3.73GGGDD47 pKa = 3.65IDD49 pKa = 6.32DD50 pKa = 5.3FDD52 pKa = 6.72PDD54 pKa = 3.83VIRR57 pKa = 11.84QSGCLEE63 pKa = 4.22KK64 pKa = 11.13GSVASANIEE73 pKa = 4.33NNTSWYY79 pKa = 9.78VFRR82 pKa = 11.84AGTGGQVGFDD92 pKa = 3.42IEE94 pKa = 4.45ALSDD98 pKa = 3.27TAEE101 pKa = 4.0WDD103 pKa = 3.62FALYY107 pKa = 10.5GPDD110 pKa = 3.83VDD112 pKa = 5.97CGDD115 pKa = 3.91ISNGTAQPIRR125 pKa = 11.84CNYY128 pKa = 8.15EE129 pKa = 3.76VNDD132 pKa = 3.71TRR134 pKa = 11.84FTGVGVNPEE143 pKa = 4.02NGQAGQPFVKK153 pKa = 10.48GSQNTYY159 pKa = 10.82DD160 pKa = 3.21EE161 pKa = 5.16WIDD164 pKa = 3.66VQPGEE169 pKa = 4.7IYY171 pKa = 10.83YY172 pKa = 10.82LLINNYY178 pKa = 7.43NTNFDD183 pKa = 4.4GDD185 pKa = 4.2PEE187 pKa = 4.72PYY189 pKa = 10.57SLTFTGNSVDD199 pKa = 4.05ADD201 pKa = 3.31QDD203 pKa = 3.66NALDD207 pKa = 3.68CTLRR211 pKa = 11.84DD212 pKa = 3.55EE213 pKa = 5.22FLGLDD218 pKa = 3.54IVACEE223 pKa = 4.1GDD225 pKa = 3.61PDD227 pKa = 3.97IVLSALNSPAGPDD240 pKa = 3.07IANVTWSVDD249 pKa = 3.54YY250 pKa = 11.19EE251 pKa = 4.36DD252 pKa = 5.73DD253 pKa = 4.42GVIDD257 pKa = 4.09AQLADD262 pKa = 4.17GPGEE266 pKa = 4.18TEE268 pKa = 4.02FTVVSPISGRR278 pKa = 11.84YY279 pKa = 7.43YY280 pKa = 11.07VEE282 pKa = 3.8ILTTLATTITDD293 pKa = 4.65DD294 pKa = 5.89ILITWYY300 pKa = 10.27GVPVLDD306 pKa = 5.39RR307 pKa = 11.84VDD309 pKa = 3.81ILDD312 pKa = 5.41DD313 pKa = 5.69LSDD316 pKa = 3.53QNNIQVFVQGDD327 pKa = 3.6GDD329 pKa = 4.16YY330 pKa = 11.45EE331 pKa = 3.93FAINNGPFQDD341 pKa = 4.19DD342 pKa = 4.25SIFRR346 pKa = 11.84DD347 pKa = 3.94VPPGINTLIINDD359 pKa = 3.81KK360 pKa = 9.92NGCGTTEE367 pKa = 4.52PIEE370 pKa = 4.07FLVVGYY376 pKa = 9.34PKK378 pKa = 10.75FFTPNNDD385 pKa = 3.17TFNDD389 pKa = 3.21TWQVKK394 pKa = 10.18GIEE397 pKa = 4.13TLIDD401 pKa = 3.4PVVFIFDD408 pKa = 3.99RR409 pKa = 11.84YY410 pKa = 10.64GKK412 pKa = 9.9LLKK415 pKa = 10.74QIDD418 pKa = 4.21EE419 pKa = 4.68TSLGWDD425 pKa = 3.34GSFNGRR431 pKa = 11.84PMPASDD437 pKa = 2.97YY438 pKa = 10.13WFRR441 pKa = 11.84LEE443 pKa = 3.92YY444 pKa = 10.89SRR446 pKa = 11.84DD447 pKa = 3.15EE448 pKa = 4.33SGIVVANTIRR458 pKa = 11.84AHH460 pKa = 5.41FTLKK464 pKa = 10.46RR465 pKa = 3.52
MM1 pKa = 6.99TRR3 pKa = 11.84NLVFLLAFLWATYY16 pKa = 10.57SLQAQNSPDD25 pKa = 3.77CRR27 pKa = 11.84SAIPVCADD35 pKa = 3.2APILNFADD43 pKa = 3.73GGGDD47 pKa = 3.65IDD49 pKa = 6.32DD50 pKa = 5.3FDD52 pKa = 6.72PDD54 pKa = 3.83VIRR57 pKa = 11.84QSGCLEE63 pKa = 4.22KK64 pKa = 11.13GSVASANIEE73 pKa = 4.33NNTSWYY79 pKa = 9.78VFRR82 pKa = 11.84AGTGGQVGFDD92 pKa = 3.42IEE94 pKa = 4.45ALSDD98 pKa = 3.27TAEE101 pKa = 4.0WDD103 pKa = 3.62FALYY107 pKa = 10.5GPDD110 pKa = 3.83VDD112 pKa = 5.97CGDD115 pKa = 3.91ISNGTAQPIRR125 pKa = 11.84CNYY128 pKa = 8.15EE129 pKa = 3.76VNDD132 pKa = 3.71TRR134 pKa = 11.84FTGVGVNPEE143 pKa = 4.02NGQAGQPFVKK153 pKa = 10.48GSQNTYY159 pKa = 10.82DD160 pKa = 3.21EE161 pKa = 5.16WIDD164 pKa = 3.66VQPGEE169 pKa = 4.7IYY171 pKa = 10.83YY172 pKa = 10.82LLINNYY178 pKa = 7.43NTNFDD183 pKa = 4.4GDD185 pKa = 4.2PEE187 pKa = 4.72PYY189 pKa = 10.57SLTFTGNSVDD199 pKa = 4.05ADD201 pKa = 3.31QDD203 pKa = 3.66NALDD207 pKa = 3.68CTLRR211 pKa = 11.84DD212 pKa = 3.55EE213 pKa = 5.22FLGLDD218 pKa = 3.54IVACEE223 pKa = 4.1GDD225 pKa = 3.61PDD227 pKa = 3.97IVLSALNSPAGPDD240 pKa = 3.07IANVTWSVDD249 pKa = 3.54YY250 pKa = 11.19EE251 pKa = 4.36DD252 pKa = 5.73DD253 pKa = 4.42GVIDD257 pKa = 4.09AQLADD262 pKa = 4.17GPGEE266 pKa = 4.18TEE268 pKa = 4.02FTVVSPISGRR278 pKa = 11.84YY279 pKa = 7.43YY280 pKa = 11.07VEE282 pKa = 3.8ILTTLATTITDD293 pKa = 4.65DD294 pKa = 5.89ILITWYY300 pKa = 10.27GVPVLDD306 pKa = 5.39RR307 pKa = 11.84VDD309 pKa = 3.81ILDD312 pKa = 5.41DD313 pKa = 5.69LSDD316 pKa = 3.53QNNIQVFVQGDD327 pKa = 3.6GDD329 pKa = 4.16YY330 pKa = 11.45EE331 pKa = 3.93FAINNGPFQDD341 pKa = 4.19DD342 pKa = 4.25SIFRR346 pKa = 11.84DD347 pKa = 3.94VPPGINTLIINDD359 pKa = 3.81KK360 pKa = 9.92NGCGTTEE367 pKa = 4.52PIEE370 pKa = 4.07FLVVGYY376 pKa = 9.34PKK378 pKa = 10.75FFTPNNDD385 pKa = 3.17TFNDD389 pKa = 3.21TWQVKK394 pKa = 10.18GIEE397 pKa = 4.13TLIDD401 pKa = 3.4PVVFIFDD408 pKa = 3.99RR409 pKa = 11.84YY410 pKa = 10.64GKK412 pKa = 9.9LLKK415 pKa = 10.74QIDD418 pKa = 4.21EE419 pKa = 4.68TSLGWDD425 pKa = 3.34GSFNGRR431 pKa = 11.84PMPASDD437 pKa = 2.97YY438 pKa = 10.13WFRR441 pKa = 11.84LEE443 pKa = 3.92YY444 pKa = 10.89SRR446 pKa = 11.84DD447 pKa = 3.15EE448 pKa = 4.33SGIVVANTIRR458 pKa = 11.84AHH460 pKa = 5.41FTLKK464 pKa = 10.46RR465 pKa = 3.52
Molecular weight: 51.22 kDa
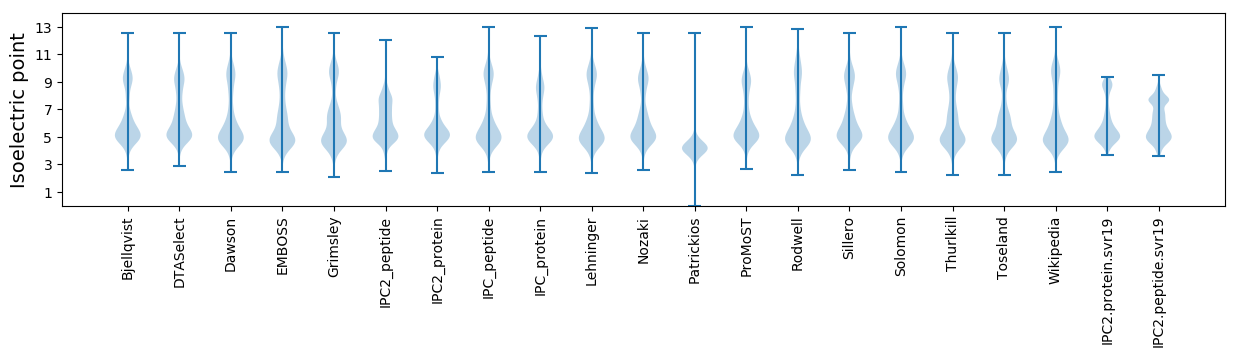
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6FMU4|A0A1I6FMU4_9FLAO DUF4328 domain-containing protein OS=Robiginitalea myxolifaciens OX=400055 GN=SAMN04490243_0087 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.89KK51 pKa = 10.65RR52 pKa = 3.21
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.89KK51 pKa = 10.65RR52 pKa = 3.21
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
998853 |
31 |
4472 |
351.2 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.924 ± 0.044 | 0.728 ± 0.015 |
5.703 ± 0.051 | 6.956 ± 0.052 |
4.821 ± 0.033 | 7.533 ± 0.052 |
1.799 ± 0.023 | 6.443 ± 0.033 |
5.152 ± 0.058 | 10.182 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.024 | 4.554 ± 0.041 |
4.218 ± 0.034 | 3.801 ± 0.027 |
5.063 ± 0.044 | 6.231 ± 0.037 |
5.393 ± 0.06 | 6.278 ± 0.039 |
1.22 ± 0.018 | 3.724 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
