
Zucchini green mottle mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
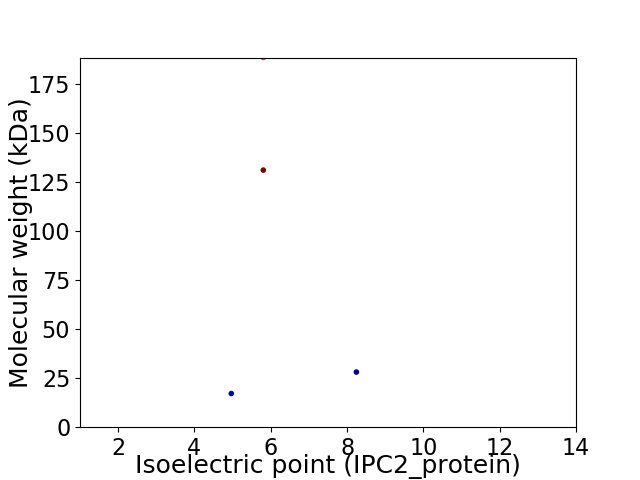
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q911R3|Q911R3_9VIRU Isoform of Q911R4 131 kDa replicase OS=Zucchini green mottle mosaic virus OX=111418 GN=replicase PE=4 SV=1
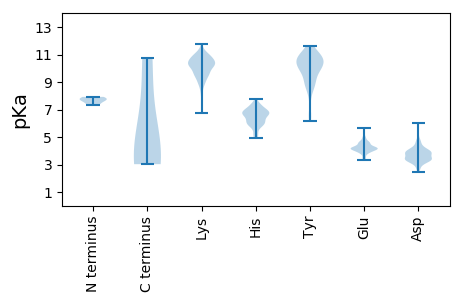
MM1 pKa = 7.92PYY3 pKa = 8.35STSGIRR9 pKa = 11.84SLPAFSKK16 pKa = 11.02SFFPYY21 pKa = 10.64LEE23 pKa = 4.89LYY25 pKa = 10.37NLLITNQGAALQTQNGKK42 pKa = 10.39DD43 pKa = 3.3ILRR46 pKa = 11.84EE47 pKa = 4.02SLVGLLSSVASPTSQFPSGVFYY69 pKa = 10.62VWSRR73 pKa = 11.84EE74 pKa = 3.84SRR76 pKa = 11.84IAALIDD82 pKa = 3.7SLFGALDD89 pKa = 3.35SRR91 pKa = 11.84NRR93 pKa = 11.84AIEE96 pKa = 3.87VEE98 pKa = 4.25NPSNPSTGEE107 pKa = 3.45ALNAVKK113 pKa = 10.64RR114 pKa = 11.84NDD116 pKa = 3.89DD117 pKa = 3.5ASTAAHH123 pKa = 6.6NDD125 pKa = 2.72IPQILSALNEE135 pKa = 4.14GAGVFDD141 pKa = 3.78RR142 pKa = 11.84ASFEE146 pKa = 4.56SAFGLVWTAGSSTSSS161 pKa = 3.03
MM1 pKa = 7.92PYY3 pKa = 8.35STSGIRR9 pKa = 11.84SLPAFSKK16 pKa = 11.02SFFPYY21 pKa = 10.64LEE23 pKa = 4.89LYY25 pKa = 10.37NLLITNQGAALQTQNGKK42 pKa = 10.39DD43 pKa = 3.3ILRR46 pKa = 11.84EE47 pKa = 4.02SLVGLLSSVASPTSQFPSGVFYY69 pKa = 10.62VWSRR73 pKa = 11.84EE74 pKa = 3.84SRR76 pKa = 11.84IAALIDD82 pKa = 3.7SLFGALDD89 pKa = 3.35SRR91 pKa = 11.84NRR93 pKa = 11.84AIEE96 pKa = 3.87VEE98 pKa = 4.25NPSNPSTGEE107 pKa = 3.45ALNAVKK113 pKa = 10.64RR114 pKa = 11.84NDD116 pKa = 3.89DD117 pKa = 3.5ASTAAHH123 pKa = 6.6NDD125 pKa = 2.72IPQILSALNEE135 pKa = 4.14GAGVFDD141 pKa = 3.78RR142 pKa = 11.84ASFEE146 pKa = 4.56SAFGLVWTAGSSTSSS161 pKa = 3.03
Molecular weight: 17.18 kDa
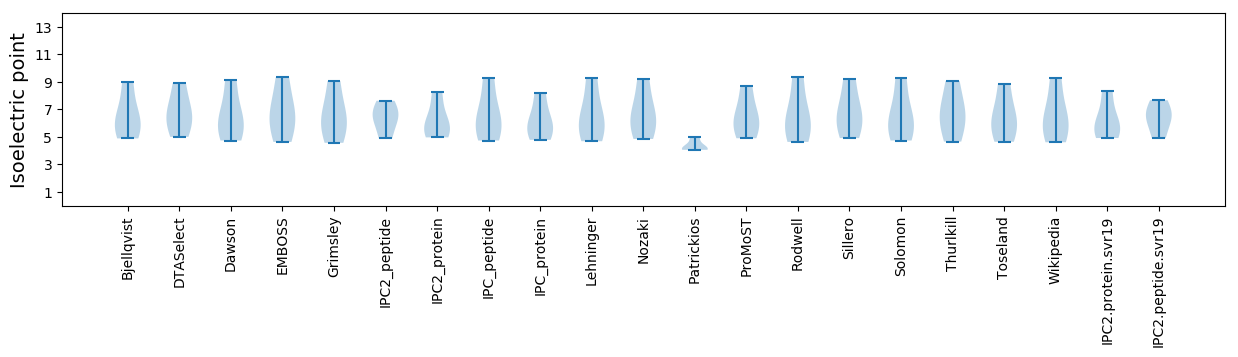
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q911R4|Q911R4_9VIRU Methyltransferase/RNA helicase OS=Zucchini green mottle mosaic virus OX=111418 GN=replicase PE=4 SV=1
MM1 pKa = 7.34SVSKK5 pKa = 11.03LGVKK9 pKa = 9.91NVLKK13 pKa = 9.88PQEE16 pKa = 4.17FVKK19 pKa = 10.84LNWVDD24 pKa = 3.84KK25 pKa = 10.35VLPDD29 pKa = 3.29MFTVFRR35 pKa = 11.84YY36 pKa = 10.24LSVTDD41 pKa = 3.3YY42 pKa = 11.59SVIKK46 pKa = 10.86SRR48 pKa = 11.84DD49 pKa = 3.78SEE51 pKa = 4.39CLIPVDD57 pKa = 4.3LLRR60 pKa = 11.84GVDD63 pKa = 3.87LSKK66 pKa = 10.97SKK68 pKa = 10.94YY69 pKa = 6.73VTLVGVVISGVWTIPEE85 pKa = 4.03NCAGGATVALVDD97 pKa = 3.8TRR99 pKa = 11.84MSMVDD104 pKa = 3.27EE105 pKa = 4.13GTICKK110 pKa = 10.41FSVAASARR118 pKa = 11.84DD119 pKa = 3.72FMVKK123 pKa = 10.45LIPNYY128 pKa = 10.46YY129 pKa = 8.55VTATDD134 pKa = 3.9ASSKK138 pKa = 9.08PWSIFVRR145 pKa = 11.84VSGVRR150 pKa = 11.84IKK152 pKa = 10.89EE153 pKa = 4.11GFSPLTLEE161 pKa = 4.2IASLVATTNSILKK174 pKa = 9.83KK175 pKa = 9.05GLRR178 pKa = 11.84VSVLEE183 pKa = 4.18SVVGSDD189 pKa = 3.59ASVNLEE195 pKa = 3.98SASEE199 pKa = 4.07KK200 pKa = 10.0VQPFFDD206 pKa = 4.12SVPITAAVISRR217 pKa = 11.84DD218 pKa = 3.03RR219 pKa = 11.84SYY221 pKa = 11.66VSKK224 pKa = 10.93SGFNRR229 pKa = 11.84AVRR232 pKa = 11.84SKK234 pKa = 10.31PPSKK238 pKa = 10.44GGKK241 pKa = 9.54KK242 pKa = 10.29FGDD245 pKa = 3.78SAEE248 pKa = 4.29SLSEE252 pKa = 4.16DD253 pKa = 3.93SASEE257 pKa = 4.22LPGLL261 pKa = 4.37
MM1 pKa = 7.34SVSKK5 pKa = 11.03LGVKK9 pKa = 9.91NVLKK13 pKa = 9.88PQEE16 pKa = 4.17FVKK19 pKa = 10.84LNWVDD24 pKa = 3.84KK25 pKa = 10.35VLPDD29 pKa = 3.29MFTVFRR35 pKa = 11.84YY36 pKa = 10.24LSVTDD41 pKa = 3.3YY42 pKa = 11.59SVIKK46 pKa = 10.86SRR48 pKa = 11.84DD49 pKa = 3.78SEE51 pKa = 4.39CLIPVDD57 pKa = 4.3LLRR60 pKa = 11.84GVDD63 pKa = 3.87LSKK66 pKa = 10.97SKK68 pKa = 10.94YY69 pKa = 6.73VTLVGVVISGVWTIPEE85 pKa = 4.03NCAGGATVALVDD97 pKa = 3.8TRR99 pKa = 11.84MSMVDD104 pKa = 3.27EE105 pKa = 4.13GTICKK110 pKa = 10.41FSVAASARR118 pKa = 11.84DD119 pKa = 3.72FMVKK123 pKa = 10.45LIPNYY128 pKa = 10.46YY129 pKa = 8.55VTATDD134 pKa = 3.9ASSKK138 pKa = 9.08PWSIFVRR145 pKa = 11.84VSGVRR150 pKa = 11.84IKK152 pKa = 10.89EE153 pKa = 4.11GFSPLTLEE161 pKa = 4.2IASLVATTNSILKK174 pKa = 9.83KK175 pKa = 9.05GLRR178 pKa = 11.84VSVLEE183 pKa = 4.18SVVGSDD189 pKa = 3.59ASVNLEE195 pKa = 3.98SASEE199 pKa = 4.07KK200 pKa = 10.0VQPFFDD206 pKa = 4.12SVPITAAVISRR217 pKa = 11.84DD218 pKa = 3.03RR219 pKa = 11.84SYY221 pKa = 11.66VSKK224 pKa = 10.93SGFNRR229 pKa = 11.84AVRR232 pKa = 11.84SKK234 pKa = 10.31PPSKK238 pKa = 10.44GGKK241 pKa = 9.54KK242 pKa = 10.29FGDD245 pKa = 3.78SAEE248 pKa = 4.29SLSEE252 pKa = 4.16DD253 pKa = 3.93SASEE257 pKa = 4.22LPGLL261 pKa = 4.37
Molecular weight: 28.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3258 |
161 |
1670 |
814.5 |
91.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.366 ± 0.435 | 1.995 ± 0.338 |
6.108 ± 0.254 | 5.648 ± 0.228 |
5.586 ± 0.337 | 4.665 ± 0.457 |
2.271 ± 0.501 | 5.218 ± 0.119 |
6.077 ± 0.458 | 8.257 ± 0.354 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.995 ± 0.177 | 3.591 ± 0.284 |
3.959 ± 0.204 | 3.376 ± 0.439 |
4.696 ± 0.069 | 8.84 ± 1.465 |
6.476 ± 0.407 | 9.269 ± 0.696 |
1.197 ± 0.007 | 3.376 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
