
Rice hoja blanca tenuivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Tenuivirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
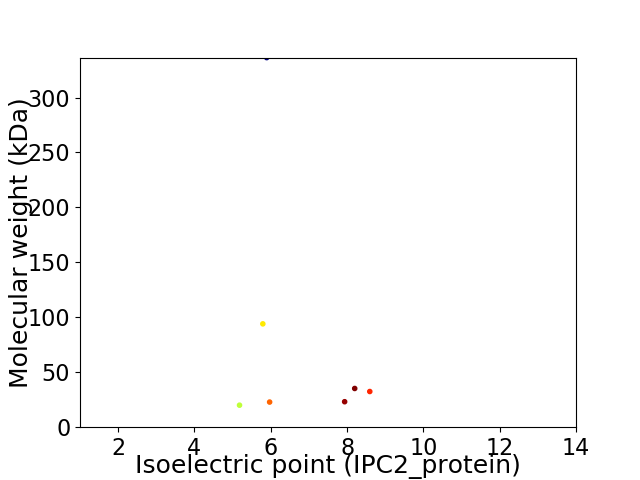
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5CMS9|A0A2H5CMS9_9VIRU NS3 protein OS=Rice hoja blanca tenuivirus OX=12332 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.8FLKK5 pKa = 10.2TDD7 pKa = 3.35VSVGPIEE14 pKa = 4.09GLNYY18 pKa = 9.71RR19 pKa = 11.84RR20 pKa = 11.84LYY22 pKa = 10.73DD23 pKa = 3.42ILPNKK28 pKa = 10.0VSDD31 pKa = 4.57NITLPDD37 pKa = 4.28LKK39 pKa = 11.15NPDD42 pKa = 3.55KK43 pKa = 10.87VTEE46 pKa = 4.16EE47 pKa = 3.89NKK49 pKa = 10.68KK50 pKa = 10.84LILCGFIYY58 pKa = 10.39VAYY61 pKa = 9.2HH62 pKa = 6.18HH63 pKa = 7.17PIEE66 pKa = 4.48TDD68 pKa = 3.1PDD70 pKa = 3.6FTSVHH75 pKa = 5.02KK76 pKa = 10.29HH77 pKa = 4.52MPGISEE83 pKa = 4.3SFLEE87 pKa = 4.36HH88 pKa = 7.3LLGTDD93 pKa = 3.58EE94 pKa = 4.74SNNTIDD100 pKa = 4.6LGKK103 pKa = 10.64LFDD106 pKa = 4.05ILQEE110 pKa = 4.2RR111 pKa = 11.84LGDD114 pKa = 4.18WITMNFLKK122 pKa = 10.71HH123 pKa = 5.63NNRR126 pKa = 11.84MSKK129 pKa = 10.49DD130 pKa = 3.53QIKK133 pKa = 8.01TLCEE137 pKa = 4.15TIVDD141 pKa = 4.29LAKK144 pKa = 10.91AEE146 pKa = 4.39GGDD149 pKa = 3.66TEE151 pKa = 4.71IYY153 pKa = 8.75EE154 pKa = 4.55AVWKK158 pKa = 10.6KK159 pKa = 9.58MPAYY163 pKa = 10.47YY164 pKa = 10.31SILLQQILHH173 pKa = 6.51KK174 pKa = 10.88
MM1 pKa = 8.12DD2 pKa = 4.8FLKK5 pKa = 10.2TDD7 pKa = 3.35VSVGPIEE14 pKa = 4.09GLNYY18 pKa = 9.71RR19 pKa = 11.84RR20 pKa = 11.84LYY22 pKa = 10.73DD23 pKa = 3.42ILPNKK28 pKa = 10.0VSDD31 pKa = 4.57NITLPDD37 pKa = 4.28LKK39 pKa = 11.15NPDD42 pKa = 3.55KK43 pKa = 10.87VTEE46 pKa = 4.16EE47 pKa = 3.89NKK49 pKa = 10.68KK50 pKa = 10.84LILCGFIYY58 pKa = 10.39VAYY61 pKa = 9.2HH62 pKa = 6.18HH63 pKa = 7.17PIEE66 pKa = 4.48TDD68 pKa = 3.1PDD70 pKa = 3.6FTSVHH75 pKa = 5.02KK76 pKa = 10.29HH77 pKa = 4.52MPGISEE83 pKa = 4.3SFLEE87 pKa = 4.36HH88 pKa = 7.3LLGTDD93 pKa = 3.58EE94 pKa = 4.74SNNTIDD100 pKa = 4.6LGKK103 pKa = 10.64LFDD106 pKa = 4.05ILQEE110 pKa = 4.2RR111 pKa = 11.84LGDD114 pKa = 4.18WITMNFLKK122 pKa = 10.71HH123 pKa = 5.63NNRR126 pKa = 11.84MSKK129 pKa = 10.49DD130 pKa = 3.53QIKK133 pKa = 8.01TLCEE137 pKa = 4.15TIVDD141 pKa = 4.29LAKK144 pKa = 10.91AEE146 pKa = 4.39GGDD149 pKa = 3.66TEE151 pKa = 4.71IYY153 pKa = 8.75EE154 pKa = 4.55AVWKK158 pKa = 10.6KK159 pKa = 9.58MPAYY163 pKa = 10.47YY164 pKa = 10.31SILLQQILHH173 pKa = 6.51KK174 pKa = 10.88
Molecular weight: 20.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q08349|Q08349_9VIRU Movement protein NS4 OS=Rice hoja blanca tenuivirus OX=12332 GN=NS4vc PE=4 SV=1
MM1 pKa = 7.49SISRR5 pKa = 11.84YY6 pKa = 9.93NPFKK10 pKa = 10.8KK11 pKa = 10.42SVILTDD17 pKa = 4.08DD18 pKa = 3.46LSEE21 pKa = 4.01RR22 pKa = 11.84AAEE25 pKa = 4.16KK26 pKa = 10.66FEE28 pKa = 4.35KK29 pKa = 10.28KK30 pKa = 9.84NKK32 pKa = 9.82RR33 pKa = 11.84KK34 pKa = 9.3LALKK38 pKa = 10.0NRR40 pKa = 11.84PLTKK44 pKa = 10.6GRR46 pKa = 11.84MTIDD50 pKa = 2.83AAATVLGLEE59 pKa = 4.45PFSFADD65 pKa = 3.58VRR67 pKa = 11.84ANSYY71 pKa = 11.86DD72 pKa = 3.51MFVAKK77 pKa = 9.87QDD79 pKa = 3.61YY80 pKa = 9.58SVCANRR86 pKa = 11.84RR87 pKa = 11.84THH89 pKa = 5.79FTIDD93 pKa = 3.17SSPLFFRR100 pKa = 11.84KK101 pKa = 9.28PLQTFPFFRR110 pKa = 11.84IATFAVIWLGIKK122 pKa = 9.91GRR124 pKa = 11.84ANGTVTFRR132 pKa = 11.84IIDD135 pKa = 3.47RR136 pKa = 11.84SYY138 pKa = 10.63TDD140 pKa = 3.49PEE142 pKa = 4.02RR143 pKa = 11.84QVEE146 pKa = 4.22VEE148 pKa = 3.5ICYY151 pKa = 10.5PMAKK155 pKa = 9.0TFAVLGSLPNFMSYY169 pKa = 10.63EE170 pKa = 4.35DD171 pKa = 3.95ADD173 pKa = 3.73KK174 pKa = 10.08MQVEE178 pKa = 4.57IVIKK182 pKa = 10.44DD183 pKa = 3.71DD184 pKa = 3.76SVQNCIISRR193 pKa = 11.84SLWFWGIEE201 pKa = 3.93RR202 pKa = 11.84TDD204 pKa = 3.94FPVPMEE210 pKa = 4.12SQKK213 pKa = 9.62TVMFEE218 pKa = 4.16FEE220 pKa = 4.63PLPDD224 pKa = 2.95RR225 pKa = 11.84TVNHH229 pKa = 6.73LSKK232 pKa = 10.77FKK234 pKa = 11.17NFTTDD239 pKa = 2.74VVQRR243 pKa = 11.84AVTTAFTTRR252 pKa = 11.84EE253 pKa = 4.02ALEE256 pKa = 4.57DD257 pKa = 3.46KK258 pKa = 10.64PGIEE262 pKa = 4.35FGVVKK267 pKa = 10.62QPGVPLVQKK276 pKa = 10.0KK277 pKa = 9.21RR278 pKa = 11.84VMIEE282 pKa = 3.52AA283 pKa = 4.29
MM1 pKa = 7.49SISRR5 pKa = 11.84YY6 pKa = 9.93NPFKK10 pKa = 10.8KK11 pKa = 10.42SVILTDD17 pKa = 4.08DD18 pKa = 3.46LSEE21 pKa = 4.01RR22 pKa = 11.84AAEE25 pKa = 4.16KK26 pKa = 10.66FEE28 pKa = 4.35KK29 pKa = 10.28KK30 pKa = 9.84NKK32 pKa = 9.82RR33 pKa = 11.84KK34 pKa = 9.3LALKK38 pKa = 10.0NRR40 pKa = 11.84PLTKK44 pKa = 10.6GRR46 pKa = 11.84MTIDD50 pKa = 2.83AAATVLGLEE59 pKa = 4.45PFSFADD65 pKa = 3.58VRR67 pKa = 11.84ANSYY71 pKa = 11.86DD72 pKa = 3.51MFVAKK77 pKa = 9.87QDD79 pKa = 3.61YY80 pKa = 9.58SVCANRR86 pKa = 11.84RR87 pKa = 11.84THH89 pKa = 5.79FTIDD93 pKa = 3.17SSPLFFRR100 pKa = 11.84KK101 pKa = 9.28PLQTFPFFRR110 pKa = 11.84IATFAVIWLGIKK122 pKa = 9.91GRR124 pKa = 11.84ANGTVTFRR132 pKa = 11.84IIDD135 pKa = 3.47RR136 pKa = 11.84SYY138 pKa = 10.63TDD140 pKa = 3.49PEE142 pKa = 4.02RR143 pKa = 11.84QVEE146 pKa = 4.22VEE148 pKa = 3.5ICYY151 pKa = 10.5PMAKK155 pKa = 9.0TFAVLGSLPNFMSYY169 pKa = 10.63EE170 pKa = 4.35DD171 pKa = 3.95ADD173 pKa = 3.73KK174 pKa = 10.08MQVEE178 pKa = 4.57IVIKK182 pKa = 10.44DD183 pKa = 3.71DD184 pKa = 3.76SVQNCIISRR193 pKa = 11.84SLWFWGIEE201 pKa = 3.93RR202 pKa = 11.84TDD204 pKa = 3.94FPVPMEE210 pKa = 4.12SQKK213 pKa = 9.62TVMFEE218 pKa = 4.16FEE220 pKa = 4.63PLPDD224 pKa = 2.95RR225 pKa = 11.84TVNHH229 pKa = 6.73LSKK232 pKa = 10.77FKK234 pKa = 11.17NFTTDD239 pKa = 2.74VVQRR243 pKa = 11.84AVTTAFTTRR252 pKa = 11.84EE253 pKa = 4.02ALEE256 pKa = 4.57DD257 pKa = 3.46KK258 pKa = 10.64PGIEE262 pKa = 4.35FGVVKK267 pKa = 10.62QPGVPLVQKK276 pKa = 10.0KK277 pKa = 9.21RR278 pKa = 11.84VMIEE282 pKa = 3.52AA283 pKa = 4.29
Molecular weight: 32.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4931 |
174 |
2918 |
704.4 |
80.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.847 ± 0.474 | 1.987 ± 0.822 |
6.469 ± 0.167 | 6.388 ± 0.6 |
4.502 ± 0.372 | 4.279 ± 0.454 |
2.373 ± 0.306 | 6.814 ± 0.588 |
7.869 ± 0.387 | 9.592 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.022 ± 0.182 | 4.786 ± 0.398 |
3.955 ± 0.533 | 2.88 ± 0.384 |
4.421 ± 0.568 | 7.747 ± 0.586 |
6.246 ± 0.624 | 6.591 ± 0.557 |
1.034 ± 0.129 | 4.198 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
