
Ying Kou virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses; Negevirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
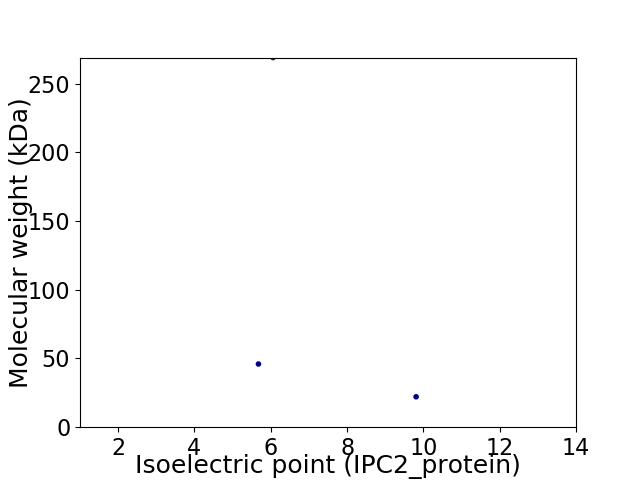
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3G1Z2Y5|A0A3G1Z2Y5_9VIRU Uncharacterized protein OS=Ying Kou virus OX=2479372 GN=ORF3 PE=4 SV=1
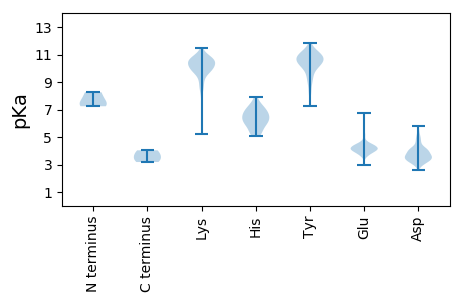
MM1 pKa = 8.26LLMILIISVVPFVRR15 pKa = 11.84SAVFVKK21 pKa = 10.6DD22 pKa = 3.19VTAQLQNTRR31 pKa = 11.84EE32 pKa = 3.84RR33 pKa = 11.84LILYY37 pKa = 9.57SKK39 pKa = 10.32IKK41 pKa = 9.64PRR43 pKa = 11.84SHH45 pKa = 6.8FVGEE49 pKa = 4.22YY50 pKa = 10.71DD51 pKa = 3.39VTPYY55 pKa = 10.89SFEE58 pKa = 4.16KK59 pKa = 10.42EE60 pKa = 4.03CMSIYY65 pKa = 9.38RR66 pKa = 11.84TNDD69 pKa = 2.57WLFANCEE76 pKa = 4.16LPAHH80 pKa = 6.94CSTPLVSKK88 pKa = 10.65INKK91 pKa = 8.29NWLGQEE97 pKa = 3.54IVLCHH102 pKa = 5.83GVHH105 pKa = 6.28PVVDD109 pKa = 4.18TPRR112 pKa = 11.84LEE114 pKa = 4.11FFTVDD119 pKa = 4.7FIPQTFSFGRR129 pKa = 11.84PISFEE134 pKa = 3.83FGKK137 pKa = 10.92VSNFTNFQPVSFFEE151 pKa = 3.77WFYY154 pKa = 11.22VVRR157 pKa = 11.84GPDD160 pKa = 3.19GYY162 pKa = 11.53GIVPKK167 pKa = 10.39FCSEE171 pKa = 3.92TFRR174 pKa = 11.84NSTALPPNVEE184 pKa = 3.82IHH186 pKa = 6.78SDD188 pKa = 3.68GDD190 pKa = 4.09SLCLRR195 pKa = 11.84RR196 pKa = 11.84VLFDD200 pKa = 4.99KK201 pKa = 10.89NDD203 pKa = 3.59CRR205 pKa = 11.84PTFYY209 pKa = 11.1EE210 pKa = 4.43SFTGLLFKK218 pKa = 11.43YY219 pKa = 9.22NFPVQFDD226 pKa = 4.08GAPFSQGAYY235 pKa = 10.03ASTNNKK241 pKa = 9.47VIGADD246 pKa = 3.28HH247 pKa = 6.16YY248 pKa = 7.73TTSNSGKK255 pKa = 10.07LPTHH259 pKa = 7.15IIDD262 pKa = 3.55KK263 pKa = 10.1VGRR266 pKa = 11.84SYY268 pKa = 10.78MVTYY272 pKa = 10.66SSYY275 pKa = 11.17EE276 pKa = 3.98DD277 pKa = 4.13PEE279 pKa = 4.61PIYY282 pKa = 10.12IVKK285 pKa = 8.72ATQVVPFEE293 pKa = 5.14DD294 pKa = 4.4SEE296 pKa = 4.53CFPIVSKK303 pKa = 10.42YY304 pKa = 8.96QSPIEE309 pKa = 4.71HH310 pKa = 6.84ILKK313 pKa = 10.24KK314 pKa = 10.13ISNFFRR320 pKa = 11.84DD321 pKa = 3.76EE322 pKa = 3.88IEE324 pKa = 4.06YY325 pKa = 10.79LLEE328 pKa = 4.21FFLVVAEE335 pKa = 5.09KK336 pKa = 10.33IASILVAIIGEE347 pKa = 4.4LLSLITTLIPYY358 pKa = 10.37GEE360 pKa = 4.52LFYY363 pKa = 10.93TSFFLACVTYY373 pKa = 10.85VFTRR377 pKa = 11.84DD378 pKa = 3.08VLLSVFPTVSIYY390 pKa = 10.92LLRR393 pKa = 11.84IYY395 pKa = 10.63LKK397 pKa = 9.86TVINN401 pKa = 4.03
MM1 pKa = 8.26LLMILIISVVPFVRR15 pKa = 11.84SAVFVKK21 pKa = 10.6DD22 pKa = 3.19VTAQLQNTRR31 pKa = 11.84EE32 pKa = 3.84RR33 pKa = 11.84LILYY37 pKa = 9.57SKK39 pKa = 10.32IKK41 pKa = 9.64PRR43 pKa = 11.84SHH45 pKa = 6.8FVGEE49 pKa = 4.22YY50 pKa = 10.71DD51 pKa = 3.39VTPYY55 pKa = 10.89SFEE58 pKa = 4.16KK59 pKa = 10.42EE60 pKa = 4.03CMSIYY65 pKa = 9.38RR66 pKa = 11.84TNDD69 pKa = 2.57WLFANCEE76 pKa = 4.16LPAHH80 pKa = 6.94CSTPLVSKK88 pKa = 10.65INKK91 pKa = 8.29NWLGQEE97 pKa = 3.54IVLCHH102 pKa = 5.83GVHH105 pKa = 6.28PVVDD109 pKa = 4.18TPRR112 pKa = 11.84LEE114 pKa = 4.11FFTVDD119 pKa = 4.7FIPQTFSFGRR129 pKa = 11.84PISFEE134 pKa = 3.83FGKK137 pKa = 10.92VSNFTNFQPVSFFEE151 pKa = 3.77WFYY154 pKa = 11.22VVRR157 pKa = 11.84GPDD160 pKa = 3.19GYY162 pKa = 11.53GIVPKK167 pKa = 10.39FCSEE171 pKa = 3.92TFRR174 pKa = 11.84NSTALPPNVEE184 pKa = 3.82IHH186 pKa = 6.78SDD188 pKa = 3.68GDD190 pKa = 4.09SLCLRR195 pKa = 11.84RR196 pKa = 11.84VLFDD200 pKa = 4.99KK201 pKa = 10.89NDD203 pKa = 3.59CRR205 pKa = 11.84PTFYY209 pKa = 11.1EE210 pKa = 4.43SFTGLLFKK218 pKa = 11.43YY219 pKa = 9.22NFPVQFDD226 pKa = 4.08GAPFSQGAYY235 pKa = 10.03ASTNNKK241 pKa = 9.47VIGADD246 pKa = 3.28HH247 pKa = 6.16YY248 pKa = 7.73TTSNSGKK255 pKa = 10.07LPTHH259 pKa = 7.15IIDD262 pKa = 3.55KK263 pKa = 10.1VGRR266 pKa = 11.84SYY268 pKa = 10.78MVTYY272 pKa = 10.66SSYY275 pKa = 11.17EE276 pKa = 3.98DD277 pKa = 4.13PEE279 pKa = 4.61PIYY282 pKa = 10.12IVKK285 pKa = 8.72ATQVVPFEE293 pKa = 5.14DD294 pKa = 4.4SEE296 pKa = 4.53CFPIVSKK303 pKa = 10.42YY304 pKa = 8.96QSPIEE309 pKa = 4.71HH310 pKa = 6.84ILKK313 pKa = 10.24KK314 pKa = 10.13ISNFFRR320 pKa = 11.84DD321 pKa = 3.76EE322 pKa = 3.88IEE324 pKa = 4.06YY325 pKa = 10.79LLEE328 pKa = 4.21FFLVVAEE335 pKa = 5.09KK336 pKa = 10.33IASILVAIIGEE347 pKa = 4.4LLSLITTLIPYY358 pKa = 10.37GEE360 pKa = 4.52LFYY363 pKa = 10.93TSFFLACVTYY373 pKa = 10.85VFTRR377 pKa = 11.84DD378 pKa = 3.08VLLSVFPTVSIYY390 pKa = 10.92LLRR393 pKa = 11.84IYY395 pKa = 10.63LKK397 pKa = 9.86TVINN401 pKa = 4.03
Molecular weight: 45.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G1Z324|A0A3G1Z324_9VIRU Non-structural protein 3 OS=Ying Kou virus OX=2479372 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.24SALRR5 pKa = 11.84AKK7 pKa = 10.43KK8 pKa = 9.76NSTVARR14 pKa = 11.84LAPVRR19 pKa = 11.84LAPPPARR26 pKa = 11.84VVVAKK31 pKa = 9.98KK32 pKa = 10.3AKK34 pKa = 9.34PSSPRR39 pKa = 11.84DD40 pKa = 3.25SGFDD44 pKa = 3.63LNKK47 pKa = 9.79VFSDD51 pKa = 3.65VSSTFVAGLNQPLVLLTLVLVAALVFTHH79 pKa = 6.54VNDD82 pKa = 4.81FSSGVVGKK90 pKa = 9.07WVSEE94 pKa = 4.02KK95 pKa = 10.93SATNSLALWVHH106 pKa = 5.12QNQVKK111 pKa = 9.98FLGLAIFAPAVLDD124 pKa = 3.66SPSRR128 pKa = 11.84LRR130 pKa = 11.84VPLALASVFWVMLVPQASIYY150 pKa = 10.25EE151 pKa = 4.09YY152 pKa = 10.55AIQAVALHH160 pKa = 6.32TYY162 pKa = 10.11FRR164 pKa = 11.84VKK166 pKa = 10.27LQNSRR171 pKa = 11.84ILIILSVAALYY182 pKa = 10.05FFGHH186 pKa = 6.49MSLAVPEE193 pKa = 4.3IKK195 pKa = 9.3KK196 pKa = 7.99TANSTLSSGG205 pKa = 3.57
MM1 pKa = 7.24SALRR5 pKa = 11.84AKK7 pKa = 10.43KK8 pKa = 9.76NSTVARR14 pKa = 11.84LAPVRR19 pKa = 11.84LAPPPARR26 pKa = 11.84VVVAKK31 pKa = 9.98KK32 pKa = 10.3AKK34 pKa = 9.34PSSPRR39 pKa = 11.84DD40 pKa = 3.25SGFDD44 pKa = 3.63LNKK47 pKa = 9.79VFSDD51 pKa = 3.65VSSTFVAGLNQPLVLLTLVLVAALVFTHH79 pKa = 6.54VNDD82 pKa = 4.81FSSGVVGKK90 pKa = 9.07WVSEE94 pKa = 4.02KK95 pKa = 10.93SATNSLALWVHH106 pKa = 5.12QNQVKK111 pKa = 9.98FLGLAIFAPAVLDD124 pKa = 3.66SPSRR128 pKa = 11.84LRR130 pKa = 11.84VPLALASVFWVMLVPQASIYY150 pKa = 10.25EE151 pKa = 4.09YY152 pKa = 10.55AIQAVALHH160 pKa = 6.32TYY162 pKa = 10.11FRR164 pKa = 11.84VKK166 pKa = 10.27LQNSRR171 pKa = 11.84ILIILSVAALYY182 pKa = 10.05FFGHH186 pKa = 6.49MSLAVPEE193 pKa = 4.3IKK195 pKa = 9.3KK196 pKa = 7.99TANSTLSSGG205 pKa = 3.57
Molecular weight: 22.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2963 |
205 |
2357 |
987.7 |
112.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.062 ± 1.826 | 2.092 ± 0.503 |
5.737 ± 1.099 | 5.737 ± 1.049 |
6.581 ± 0.997 | 4.387 ± 0.258 |
2.497 ± 0.26 | 5.029 ± 1.027 |
5.299 ± 0.228 | 9.112 ± 0.972 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.517 | 5.164 ± 0.606 |
4.522 ± 0.803 | 2.734 ± 0.184 |
5.872 ± 0.886 | 9.146 ± 0.548 |
5.501 ± 0.478 | 8.91 ± 1.117 |
0.54 ± 0.257 | 3.915 ± 0.615 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
