
Amphritea balenae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Amphritea
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
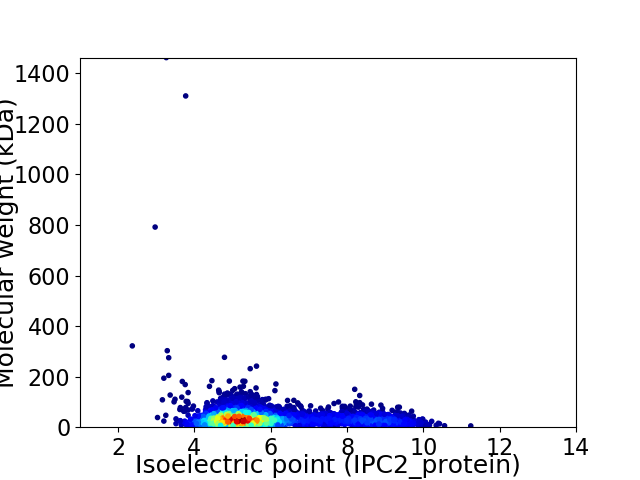
Virtual 2D-PAGE plot for 4082 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
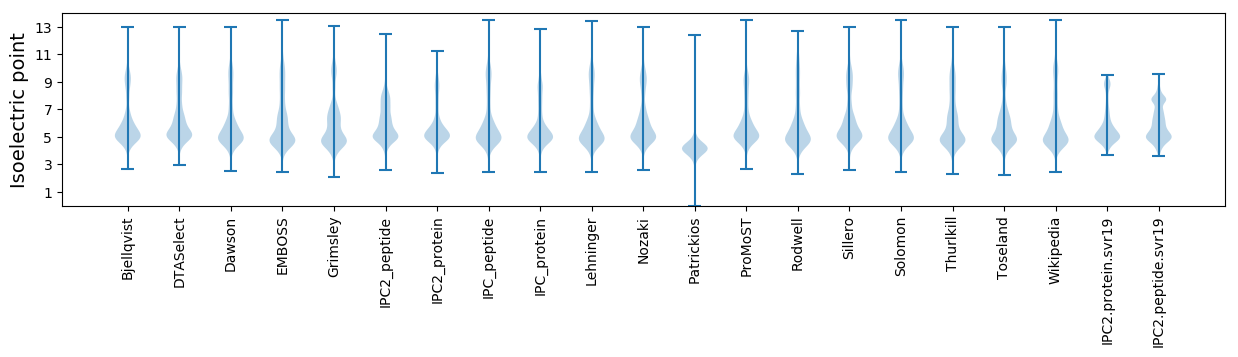
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P1SIE3|A0A3P1SIE3_9GAMM Thiosulfate reductase OS=Amphritea balenae OX=452629 GN=EHS89_19010 PE=4 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84TFSSFFKK9 pKa = 9.94LTSFIMGSVFAAALVIMMTVAAVDD33 pKa = 3.51SLGVFEE39 pKa = 6.03LDD41 pKa = 4.67GDD43 pKa = 4.18ATDD46 pKa = 4.33LASGTGSAGDD56 pKa = 4.15DD57 pKa = 3.3WSSLYY62 pKa = 10.06IDD64 pKa = 4.2CPEE67 pKa = 4.09GNVRR71 pKa = 11.84DD72 pKa = 4.29NPPGDD77 pKa = 4.05DD78 pKa = 4.33CPPGLSSVFTGIINDD93 pKa = 3.97EE94 pKa = 4.29NPSPVDD100 pKa = 3.45GTPDD104 pKa = 3.69SSIFWKK110 pKa = 10.67GGSKK114 pKa = 10.35DD115 pKa = 3.3IHH117 pKa = 7.07DD118 pKa = 4.3VPDD121 pKa = 2.71WWFKK125 pKa = 11.18AGSTPDD131 pKa = 3.36KK132 pKa = 11.1DD133 pKa = 4.08QITNAYY139 pKa = 8.37AAAYY143 pKa = 10.14SLTSGNGIHH152 pKa = 6.04QVGDD156 pKa = 3.96LIVNFGLDD164 pKa = 2.9RR165 pKa = 11.84FANNGDD171 pKa = 3.06AHH173 pKa = 6.67AGFWFFHH180 pKa = 6.34QDD182 pKa = 3.19VGPDD186 pKa = 3.33PSRR189 pKa = 11.84PSRR192 pKa = 11.84FTGMHH197 pKa = 4.2QQKK200 pKa = 10.26NISIGQRR207 pKa = 11.84GDD209 pKa = 3.03ILVLVEE215 pKa = 4.3YY216 pKa = 9.59PQGSNANPLIRR227 pKa = 11.84VYY229 pKa = 10.48EE230 pKa = 4.12WDD232 pKa = 4.27PLDD235 pKa = 4.88EE236 pKa = 6.22GDD238 pKa = 5.98DD239 pKa = 3.67NAAKK243 pKa = 10.28NLHH246 pKa = 5.56QLYY249 pKa = 10.8DD250 pKa = 3.84SDD252 pKa = 4.3TSGQSATCGVGSGGNKK268 pKa = 9.08LACAITNDD276 pKa = 3.61GSVNAPWSYY285 pKa = 10.12TPKK288 pKa = 10.21SGPGGSFPAEE298 pKa = 3.88SFYY301 pKa = 11.23EE302 pKa = 4.17GSINFTQLLGMTPCFSSFLAEE323 pKa = 3.97TRR325 pKa = 11.84ASRR328 pKa = 11.84SEE330 pKa = 3.92DD331 pKa = 3.05AKK333 pKa = 11.2LWDD336 pKa = 3.69HH337 pKa = 6.92VGGEE341 pKa = 4.52FDD343 pKa = 3.73LCSIDD348 pKa = 3.7VLKK351 pKa = 10.33TGDD354 pKa = 3.82TLSKK358 pKa = 9.97VTDD361 pKa = 3.72PVDD364 pKa = 3.41YY365 pKa = 9.93TFKK368 pKa = 9.84ITNTGAITLYY378 pKa = 10.26QDD380 pKa = 4.81FIIDD384 pKa = 3.81TLLGDD389 pKa = 4.54LVDD392 pKa = 4.59PVPASHH398 pKa = 7.19LYY400 pKa = 8.28VTSSDD405 pKa = 4.25CGPQGAALLAGHH417 pKa = 6.63YY418 pKa = 7.44CTIEE422 pKa = 3.59ATRR425 pKa = 11.84IVVGDD430 pKa = 4.05EE431 pKa = 3.6ADD433 pKa = 4.22GIYY436 pKa = 10.58DD437 pKa = 3.33SSGVLQILPNTLNVSYY453 pKa = 11.18NNQSDD458 pKa = 4.08LAGDD462 pKa = 4.09SVDD465 pKa = 4.18GSDD468 pKa = 3.71DD469 pKa = 3.47HH470 pKa = 7.94SVNLFQPSFTLVKK483 pKa = 10.0SCPIGIGSGDD493 pKa = 3.86GEE495 pKa = 4.42RR496 pKa = 11.84AWKK499 pKa = 10.39VGEE502 pKa = 4.01PLDD505 pKa = 4.78FRR507 pKa = 11.84FTLTNTSSSDD517 pKa = 3.52TPIMEE522 pKa = 5.2LGSLTDD528 pKa = 4.12PLFGGNILALLPQAKK543 pKa = 9.51QNALAEE549 pKa = 4.14LAIDD553 pKa = 3.73EE554 pKa = 5.27SIDD557 pKa = 3.58FVLSYY562 pKa = 10.39TPSSNDD568 pKa = 2.79IPGVTNTADD577 pKa = 3.25AVYY580 pKa = 10.36SPQGFTNQIAANNGTPASVFCPVALPGPATILIEE614 pKa = 4.18KK615 pKa = 8.67EE616 pKa = 3.74LRR618 pKa = 11.84GGGGVTFTFTGGGLLVPGLTPVIGGNTTEE647 pKa = 4.37SPHH650 pKa = 6.02GAVRR654 pKa = 11.84DD655 pKa = 3.82AAVDD659 pKa = 4.84GYY661 pKa = 11.34AVTGSISVSTTFGTSTQYY679 pKa = 10.74TVTEE683 pKa = 4.23SGLPAGTTFEE693 pKa = 4.6SVSCRR698 pKa = 11.84NPDD701 pKa = 3.37GEE703 pKa = 5.49LDD705 pKa = 3.75TDD707 pKa = 4.36SSASGATATINVDD720 pKa = 3.15EE721 pKa = 5.3LEE723 pKa = 4.41VVVCRR728 pKa = 11.84FINIVEE734 pKa = 4.25PKK736 pKa = 8.5LTLVKK741 pKa = 10.59SVTNDD746 pKa = 2.93NGGNAVPGDD755 pKa = 3.94FEE757 pKa = 5.26LVLNGGIYY765 pKa = 10.07SDD767 pKa = 3.99TVFSDD772 pKa = 3.34GAMPTVVAEE781 pKa = 4.05TAYY784 pKa = 9.81TLTEE788 pKa = 4.12NPFPGYY794 pKa = 10.36SQDD797 pKa = 3.51GDD799 pKa = 4.05VACVDD804 pKa = 4.35DD805 pKa = 4.47NTAQTVSHH813 pKa = 7.13PVTLDD818 pKa = 3.38FGQEE822 pKa = 3.99VTCTVVNDD830 pKa = 5.38DD831 pKa = 3.57ITPTITLIKK840 pKa = 10.35EE841 pKa = 4.21VVNDD845 pKa = 3.45NGGNAGVNDD854 pKa = 4.77FGLSIGGTGVNSGVTLDD871 pKa = 3.39VDD873 pKa = 4.04ANIAIALTEE882 pKa = 4.38AGLTGYY888 pKa = 10.0EE889 pKa = 4.36FVSIGQGGGDD899 pKa = 3.48AAKK902 pKa = 10.55CPAVLNGTVTLDD914 pKa = 3.38EE915 pKa = 5.43DD916 pKa = 3.72EE917 pKa = 6.14HH918 pKa = 5.69ITCTIVNDD926 pKa = 5.11DD927 pKa = 3.85IAPSITLIKK936 pKa = 10.35EE937 pKa = 4.16VINDD941 pKa = 3.41NGGNAGVNDD950 pKa = 4.77FGLSIGGTGVNSGEE964 pKa = 4.28TLDD967 pKa = 4.36VDD969 pKa = 4.22ANTAIALTEE978 pKa = 4.36AGLTGYY984 pKa = 10.43NFVSIGQGGGDD995 pKa = 3.6SAKK998 pKa = 10.59CPAVLNGTVTLDD1010 pKa = 3.32EE1011 pKa = 5.3DD1012 pKa = 3.86EE1013 pKa = 5.64HH1014 pKa = 5.28VTCTIVNDD1022 pKa = 5.23DD1023 pKa = 3.71ITPTITLIKK1032 pKa = 10.29EE1033 pKa = 4.23VINDD1037 pKa = 3.41NGGNAGVNDD1046 pKa = 4.77FGLSIGGTGVNSGLTLDD1063 pKa = 4.1VDD1065 pKa = 4.31ANTAIALTEE1074 pKa = 4.36AGLTGYY1080 pKa = 10.43NFVSIGQGNGDD1091 pKa = 3.83SAKK1094 pKa = 10.58CPAVLNGTVTLNEE1107 pKa = 4.7DD1108 pKa = 3.41EE1109 pKa = 5.59HH1110 pKa = 5.5VTCTIVNDD1118 pKa = 5.23DD1119 pKa = 3.71ITPTITLIKK1128 pKa = 10.39HH1129 pKa = 6.06VINDD1133 pKa = 3.45NGGNAGVNDD1142 pKa = 4.77FGLSIGGAGVNSGVTLDD1159 pKa = 3.62VEE1161 pKa = 4.55ANIAIALTEE1170 pKa = 4.06VGLTGYY1176 pKa = 9.91EE1177 pKa = 4.14FVSIGQGNNDD1187 pKa = 3.57SAKK1190 pKa = 10.53CPAVLNGTVTLDD1202 pKa = 3.32EE1203 pKa = 5.3DD1204 pKa = 3.86EE1205 pKa = 5.64HH1206 pKa = 5.28VTCTIVNDD1214 pKa = 5.23DD1215 pKa = 3.71ITPTITLIKK1224 pKa = 10.29EE1225 pKa = 4.23VINDD1229 pKa = 3.41NGGNAGVNDD1238 pKa = 4.77FGLSIGGAGVNSGLTLDD1255 pKa = 4.1VDD1257 pKa = 4.31ANTAIALTEE1266 pKa = 4.36AGLTGYY1272 pKa = 10.43NFVSIGQGNGDD1283 pKa = 3.83SAKK1286 pKa = 10.58CPAVLNGTVTLDD1298 pKa = 3.32EE1299 pKa = 5.3DD1300 pKa = 3.86EE1301 pKa = 5.64HH1302 pKa = 5.28VTCTIVNDD1310 pKa = 5.23DD1311 pKa = 3.71ITPTITLIKK1320 pKa = 10.39HH1321 pKa = 6.06VINDD1325 pKa = 3.45NGGNAGVNDD1334 pKa = 4.77FGLSIGGAGVNSGVTLDD1351 pKa = 3.39VDD1353 pKa = 4.04ANIAIALTEE1362 pKa = 4.38AGLTGYY1368 pKa = 10.0EE1369 pKa = 4.36FVSIGQGNNDD1379 pKa = 3.57SAKK1382 pKa = 10.53CPAVLNGTVTLNEE1395 pKa = 4.7DD1396 pKa = 3.41EE1397 pKa = 5.59HH1398 pKa = 5.5VTCTIVNDD1406 pKa = 5.23DD1407 pKa = 3.71ITPTITLIKK1416 pKa = 10.27EE1417 pKa = 4.24VINDD1421 pKa = 3.8DD1422 pKa = 3.77GGTAGVNDD1430 pKa = 4.82FGLSIGGTGVNSGVTLDD1447 pKa = 3.72VDD1449 pKa = 4.08ANTAIALNEE1458 pKa = 4.27AGLMGYY1464 pKa = 9.37EE1465 pKa = 4.4FVSIGQGNGDD1475 pKa = 3.83SAKK1478 pKa = 10.58CPAVLNGTVTLNEE1491 pKa = 4.7DD1492 pKa = 3.41EE1493 pKa = 5.59HH1494 pKa = 5.5VTCTIVNDD1502 pKa = 3.92DD1503 pKa = 4.82LLVEE1507 pKa = 4.91FCPEE1511 pKa = 4.11PGVSGRR1517 pKa = 11.84PKK1519 pKa = 10.11EE1520 pKa = 4.04LTMMYY1525 pKa = 10.33DD1526 pKa = 3.65GVSDD1530 pKa = 3.47RR1531 pKa = 11.84TGPQTGNEE1539 pKa = 4.12VVIDD1543 pKa = 4.09PFNVVAPDD1551 pKa = 3.42EE1552 pKa = 4.76VYY1554 pKa = 10.96VEE1556 pKa = 4.82VYY1558 pKa = 6.67THH1560 pKa = 6.9RR1561 pKa = 11.84GKK1563 pKa = 10.93LVLTTGALEE1572 pKa = 4.45EE1573 pKa = 4.78GDD1575 pKa = 3.87SFSYY1579 pKa = 10.85AGFKK1583 pKa = 10.79GGVSPRR1589 pKa = 11.84SRR1591 pKa = 11.84FLIYY1595 pKa = 10.37DD1596 pKa = 3.68YY1597 pKa = 10.95ATGDD1601 pKa = 4.62LIQTVQFHH1609 pKa = 6.18TSCSQPINLGDD1620 pKa = 3.74SYY1622 pKa = 12.14GLLTVSGGILGKK1634 pKa = 10.46
MM1 pKa = 7.47RR2 pKa = 11.84TFSSFFKK9 pKa = 9.94LTSFIMGSVFAAALVIMMTVAAVDD33 pKa = 3.51SLGVFEE39 pKa = 6.03LDD41 pKa = 4.67GDD43 pKa = 4.18ATDD46 pKa = 4.33LASGTGSAGDD56 pKa = 4.15DD57 pKa = 3.3WSSLYY62 pKa = 10.06IDD64 pKa = 4.2CPEE67 pKa = 4.09GNVRR71 pKa = 11.84DD72 pKa = 4.29NPPGDD77 pKa = 4.05DD78 pKa = 4.33CPPGLSSVFTGIINDD93 pKa = 3.97EE94 pKa = 4.29NPSPVDD100 pKa = 3.45GTPDD104 pKa = 3.69SSIFWKK110 pKa = 10.67GGSKK114 pKa = 10.35DD115 pKa = 3.3IHH117 pKa = 7.07DD118 pKa = 4.3VPDD121 pKa = 2.71WWFKK125 pKa = 11.18AGSTPDD131 pKa = 3.36KK132 pKa = 11.1DD133 pKa = 4.08QITNAYY139 pKa = 8.37AAAYY143 pKa = 10.14SLTSGNGIHH152 pKa = 6.04QVGDD156 pKa = 3.96LIVNFGLDD164 pKa = 2.9RR165 pKa = 11.84FANNGDD171 pKa = 3.06AHH173 pKa = 6.67AGFWFFHH180 pKa = 6.34QDD182 pKa = 3.19VGPDD186 pKa = 3.33PSRR189 pKa = 11.84PSRR192 pKa = 11.84FTGMHH197 pKa = 4.2QQKK200 pKa = 10.26NISIGQRR207 pKa = 11.84GDD209 pKa = 3.03ILVLVEE215 pKa = 4.3YY216 pKa = 9.59PQGSNANPLIRR227 pKa = 11.84VYY229 pKa = 10.48EE230 pKa = 4.12WDD232 pKa = 4.27PLDD235 pKa = 4.88EE236 pKa = 6.22GDD238 pKa = 5.98DD239 pKa = 3.67NAAKK243 pKa = 10.28NLHH246 pKa = 5.56QLYY249 pKa = 10.8DD250 pKa = 3.84SDD252 pKa = 4.3TSGQSATCGVGSGGNKK268 pKa = 9.08LACAITNDD276 pKa = 3.61GSVNAPWSYY285 pKa = 10.12TPKK288 pKa = 10.21SGPGGSFPAEE298 pKa = 3.88SFYY301 pKa = 11.23EE302 pKa = 4.17GSINFTQLLGMTPCFSSFLAEE323 pKa = 3.97TRR325 pKa = 11.84ASRR328 pKa = 11.84SEE330 pKa = 3.92DD331 pKa = 3.05AKK333 pKa = 11.2LWDD336 pKa = 3.69HH337 pKa = 6.92VGGEE341 pKa = 4.52FDD343 pKa = 3.73LCSIDD348 pKa = 3.7VLKK351 pKa = 10.33TGDD354 pKa = 3.82TLSKK358 pKa = 9.97VTDD361 pKa = 3.72PVDD364 pKa = 3.41YY365 pKa = 9.93TFKK368 pKa = 9.84ITNTGAITLYY378 pKa = 10.26QDD380 pKa = 4.81FIIDD384 pKa = 3.81TLLGDD389 pKa = 4.54LVDD392 pKa = 4.59PVPASHH398 pKa = 7.19LYY400 pKa = 8.28VTSSDD405 pKa = 4.25CGPQGAALLAGHH417 pKa = 6.63YY418 pKa = 7.44CTIEE422 pKa = 3.59ATRR425 pKa = 11.84IVVGDD430 pKa = 4.05EE431 pKa = 3.6ADD433 pKa = 4.22GIYY436 pKa = 10.58DD437 pKa = 3.33SSGVLQILPNTLNVSYY453 pKa = 11.18NNQSDD458 pKa = 4.08LAGDD462 pKa = 4.09SVDD465 pKa = 4.18GSDD468 pKa = 3.71DD469 pKa = 3.47HH470 pKa = 7.94SVNLFQPSFTLVKK483 pKa = 10.0SCPIGIGSGDD493 pKa = 3.86GEE495 pKa = 4.42RR496 pKa = 11.84AWKK499 pKa = 10.39VGEE502 pKa = 4.01PLDD505 pKa = 4.78FRR507 pKa = 11.84FTLTNTSSSDD517 pKa = 3.52TPIMEE522 pKa = 5.2LGSLTDD528 pKa = 4.12PLFGGNILALLPQAKK543 pKa = 9.51QNALAEE549 pKa = 4.14LAIDD553 pKa = 3.73EE554 pKa = 5.27SIDD557 pKa = 3.58FVLSYY562 pKa = 10.39TPSSNDD568 pKa = 2.79IPGVTNTADD577 pKa = 3.25AVYY580 pKa = 10.36SPQGFTNQIAANNGTPASVFCPVALPGPATILIEE614 pKa = 4.18KK615 pKa = 8.67EE616 pKa = 3.74LRR618 pKa = 11.84GGGGVTFTFTGGGLLVPGLTPVIGGNTTEE647 pKa = 4.37SPHH650 pKa = 6.02GAVRR654 pKa = 11.84DD655 pKa = 3.82AAVDD659 pKa = 4.84GYY661 pKa = 11.34AVTGSISVSTTFGTSTQYY679 pKa = 10.74TVTEE683 pKa = 4.23SGLPAGTTFEE693 pKa = 4.6SVSCRR698 pKa = 11.84NPDD701 pKa = 3.37GEE703 pKa = 5.49LDD705 pKa = 3.75TDD707 pKa = 4.36SSASGATATINVDD720 pKa = 3.15EE721 pKa = 5.3LEE723 pKa = 4.41VVVCRR728 pKa = 11.84FINIVEE734 pKa = 4.25PKK736 pKa = 8.5LTLVKK741 pKa = 10.59SVTNDD746 pKa = 2.93NGGNAVPGDD755 pKa = 3.94FEE757 pKa = 5.26LVLNGGIYY765 pKa = 10.07SDD767 pKa = 3.99TVFSDD772 pKa = 3.34GAMPTVVAEE781 pKa = 4.05TAYY784 pKa = 9.81TLTEE788 pKa = 4.12NPFPGYY794 pKa = 10.36SQDD797 pKa = 3.51GDD799 pKa = 4.05VACVDD804 pKa = 4.35DD805 pKa = 4.47NTAQTVSHH813 pKa = 7.13PVTLDD818 pKa = 3.38FGQEE822 pKa = 3.99VTCTVVNDD830 pKa = 5.38DD831 pKa = 3.57ITPTITLIKK840 pKa = 10.35EE841 pKa = 4.21VVNDD845 pKa = 3.45NGGNAGVNDD854 pKa = 4.77FGLSIGGTGVNSGVTLDD871 pKa = 3.39VDD873 pKa = 4.04ANIAIALTEE882 pKa = 4.38AGLTGYY888 pKa = 10.0EE889 pKa = 4.36FVSIGQGGGDD899 pKa = 3.48AAKK902 pKa = 10.55CPAVLNGTVTLDD914 pKa = 3.38EE915 pKa = 5.43DD916 pKa = 3.72EE917 pKa = 6.14HH918 pKa = 5.69ITCTIVNDD926 pKa = 5.11DD927 pKa = 3.85IAPSITLIKK936 pKa = 10.35EE937 pKa = 4.16VINDD941 pKa = 3.41NGGNAGVNDD950 pKa = 4.77FGLSIGGTGVNSGEE964 pKa = 4.28TLDD967 pKa = 4.36VDD969 pKa = 4.22ANTAIALTEE978 pKa = 4.36AGLTGYY984 pKa = 10.43NFVSIGQGGGDD995 pKa = 3.6SAKK998 pKa = 10.59CPAVLNGTVTLDD1010 pKa = 3.32EE1011 pKa = 5.3DD1012 pKa = 3.86EE1013 pKa = 5.64HH1014 pKa = 5.28VTCTIVNDD1022 pKa = 5.23DD1023 pKa = 3.71ITPTITLIKK1032 pKa = 10.29EE1033 pKa = 4.23VINDD1037 pKa = 3.41NGGNAGVNDD1046 pKa = 4.77FGLSIGGTGVNSGLTLDD1063 pKa = 4.1VDD1065 pKa = 4.31ANTAIALTEE1074 pKa = 4.36AGLTGYY1080 pKa = 10.43NFVSIGQGNGDD1091 pKa = 3.83SAKK1094 pKa = 10.58CPAVLNGTVTLNEE1107 pKa = 4.7DD1108 pKa = 3.41EE1109 pKa = 5.59HH1110 pKa = 5.5VTCTIVNDD1118 pKa = 5.23DD1119 pKa = 3.71ITPTITLIKK1128 pKa = 10.39HH1129 pKa = 6.06VINDD1133 pKa = 3.45NGGNAGVNDD1142 pKa = 4.77FGLSIGGAGVNSGVTLDD1159 pKa = 3.62VEE1161 pKa = 4.55ANIAIALTEE1170 pKa = 4.06VGLTGYY1176 pKa = 9.91EE1177 pKa = 4.14FVSIGQGNNDD1187 pKa = 3.57SAKK1190 pKa = 10.53CPAVLNGTVTLDD1202 pKa = 3.32EE1203 pKa = 5.3DD1204 pKa = 3.86EE1205 pKa = 5.64HH1206 pKa = 5.28VTCTIVNDD1214 pKa = 5.23DD1215 pKa = 3.71ITPTITLIKK1224 pKa = 10.29EE1225 pKa = 4.23VINDD1229 pKa = 3.41NGGNAGVNDD1238 pKa = 4.77FGLSIGGAGVNSGLTLDD1255 pKa = 4.1VDD1257 pKa = 4.31ANTAIALTEE1266 pKa = 4.36AGLTGYY1272 pKa = 10.43NFVSIGQGNGDD1283 pKa = 3.83SAKK1286 pKa = 10.58CPAVLNGTVTLDD1298 pKa = 3.32EE1299 pKa = 5.3DD1300 pKa = 3.86EE1301 pKa = 5.64HH1302 pKa = 5.28VTCTIVNDD1310 pKa = 5.23DD1311 pKa = 3.71ITPTITLIKK1320 pKa = 10.39HH1321 pKa = 6.06VINDD1325 pKa = 3.45NGGNAGVNDD1334 pKa = 4.77FGLSIGGAGVNSGVTLDD1351 pKa = 3.39VDD1353 pKa = 4.04ANIAIALTEE1362 pKa = 4.38AGLTGYY1368 pKa = 10.0EE1369 pKa = 4.36FVSIGQGNNDD1379 pKa = 3.57SAKK1382 pKa = 10.53CPAVLNGTVTLNEE1395 pKa = 4.7DD1396 pKa = 3.41EE1397 pKa = 5.59HH1398 pKa = 5.5VTCTIVNDD1406 pKa = 5.23DD1407 pKa = 3.71ITPTITLIKK1416 pKa = 10.27EE1417 pKa = 4.24VINDD1421 pKa = 3.8DD1422 pKa = 3.77GGTAGVNDD1430 pKa = 4.82FGLSIGGTGVNSGVTLDD1447 pKa = 3.72VDD1449 pKa = 4.08ANTAIALNEE1458 pKa = 4.27AGLMGYY1464 pKa = 9.37EE1465 pKa = 4.4FVSIGQGNGDD1475 pKa = 3.83SAKK1478 pKa = 10.58CPAVLNGTVTLNEE1491 pKa = 4.7DD1492 pKa = 3.41EE1493 pKa = 5.59HH1494 pKa = 5.5VTCTIVNDD1502 pKa = 3.92DD1503 pKa = 4.82LLVEE1507 pKa = 4.91FCPEE1511 pKa = 4.11PGVSGRR1517 pKa = 11.84PKK1519 pKa = 10.11EE1520 pKa = 4.04LTMMYY1525 pKa = 10.33DD1526 pKa = 3.65GVSDD1530 pKa = 3.47RR1531 pKa = 11.84TGPQTGNEE1539 pKa = 4.12VVIDD1543 pKa = 4.09PFNVVAPDD1551 pKa = 3.42EE1552 pKa = 4.76VYY1554 pKa = 10.96VEE1556 pKa = 4.82VYY1558 pKa = 6.67THH1560 pKa = 6.9RR1561 pKa = 11.84GKK1563 pKa = 10.93LVLTTGALEE1572 pKa = 4.45EE1573 pKa = 4.78GDD1575 pKa = 3.87SFSYY1579 pKa = 10.85AGFKK1583 pKa = 10.79GGVSPRR1589 pKa = 11.84SRR1591 pKa = 11.84FLIYY1595 pKa = 10.37DD1596 pKa = 3.68YY1597 pKa = 10.95ATGDD1601 pKa = 4.62LIQTVQFHH1609 pKa = 6.18TSCSQPINLGDD1620 pKa = 3.74SYY1622 pKa = 12.14GLLTVSGGILGKK1634 pKa = 10.46
Molecular weight: 168.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P1SQJ5|A0A3P1SQJ5_9GAMM Glycolate oxidase subunit GlcE OS=Amphritea balenae OX=452629 GN=glcE PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPSGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.81GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPSGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.81GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1376603 |
21 |
14333 |
337.2 |
37.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.866 ± 0.046 | 1.06 ± 0.021 |
5.896 ± 0.089 | 6.269 ± 0.051 |
3.867 ± 0.029 | 7.343 ± 0.068 |
2.074 ± 0.029 | 6.263 ± 0.031 |
4.429 ± 0.051 | 10.727 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.607 ± 0.032 | 3.953 ± 0.038 |
4.103 ± 0.03 | 4.808 ± 0.053 |
4.94 ± 0.071 | 6.624 ± 0.041 |
5.326 ± 0.117 | 6.783 ± 0.045 |
1.213 ± 0.015 | 2.849 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
