
Halovenus sp. WSH3
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halovenus; unclassified Halovenus
Average proteome isoelectric point is 4.79
Get precalculated fractions of proteins
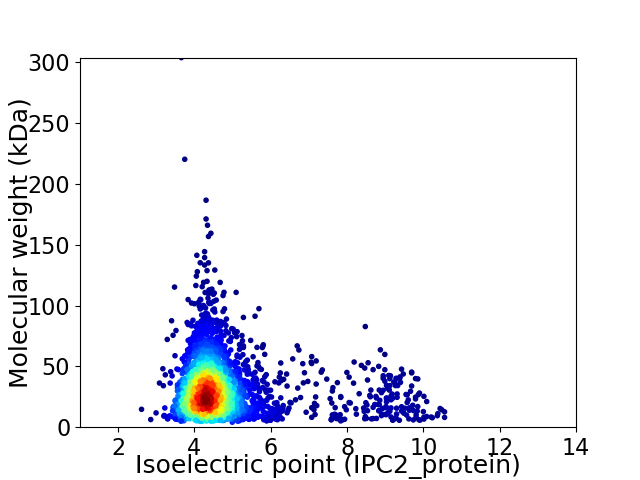
Virtual 2D-PAGE plot for 3185 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B0T6E9|A0A6B0T6E9_9EURY Winged helix-turn-helix transcriptional regulator OS=Halovenus sp. WSH3 OX=2692199 GN=GRX03_09135 PE=4 SV=1
MM1 pKa = 6.48TAEE4 pKa = 4.91RR5 pKa = 11.84SPEE8 pKa = 3.93PDD10 pKa = 3.22PQVEE14 pKa = 4.24QFLQLYY20 pKa = 10.62DD21 pKa = 3.89SMDD24 pKa = 3.44VPEE27 pKa = 4.63LHH29 pKa = 6.74EE30 pKa = 4.69LSPPEE35 pKa = 3.78ARR37 pKa = 11.84EE38 pKa = 3.86VQEE41 pKa = 4.53QLSQADD47 pKa = 3.94PDD49 pKa = 3.62IDD51 pKa = 4.08LPTVEE56 pKa = 5.73DD57 pKa = 3.83RR58 pKa = 11.84TIDD61 pKa = 3.94GPDD64 pKa = 3.18GEE66 pKa = 4.56IPIRR70 pKa = 11.84LYY72 pKa = 10.83DD73 pKa = 3.69PRR75 pKa = 11.84TEE77 pKa = 4.08EE78 pKa = 4.21TEE80 pKa = 4.04TPLILFYY87 pKa = 10.87HH88 pKa = 6.6GGGFVVGSLDD98 pKa = 3.58THH100 pKa = 7.33DD101 pKa = 4.41GPCRR105 pKa = 11.84KK106 pKa = 9.21LAAEE110 pKa = 3.91TGYY113 pKa = 10.07PVVSVDD119 pKa = 3.24YY120 pKa = 10.49RR121 pKa = 11.84LAPEE125 pKa = 4.58HH126 pKa = 6.76PFPAGLNDD134 pKa = 4.41CYY136 pKa = 11.2AALEE140 pKa = 4.37WASEE144 pKa = 3.95TDD146 pKa = 3.19EE147 pKa = 5.37LGIDD151 pKa = 3.37SGQIVVAGDD160 pKa = 3.48SAGGNFAAAVSLLSRR175 pKa = 11.84DD176 pKa = 3.22RR177 pKa = 11.84DD178 pKa = 3.85GPEE181 pKa = 3.12IAAQVLVYY189 pKa = 10.07PVTGDD194 pKa = 3.31AAEE197 pKa = 4.24TDD199 pKa = 4.17SYY201 pKa = 11.21DD202 pKa = 3.82QNGEE206 pKa = 4.25GYY208 pKa = 10.43FLTTEE213 pKa = 4.01TMEE216 pKa = 4.3WFHH219 pKa = 6.98DD220 pKa = 3.84QYY222 pKa = 11.51LDD224 pKa = 4.52DD225 pKa = 6.43PIDD228 pKa = 3.66EE229 pKa = 4.54GNIYY233 pKa = 9.7VAPRR237 pKa = 11.84LAADD241 pKa = 4.89LSDD244 pKa = 4.79LPPATVVTAGYY255 pKa = 10.56DD256 pKa = 3.49PLRR259 pKa = 11.84DD260 pKa = 3.96DD261 pKa = 3.97GALYY265 pKa = 10.82AEE267 pKa = 4.74RR268 pKa = 11.84LRR270 pKa = 11.84DD271 pKa = 3.48AGVEE275 pKa = 4.24VSHH278 pKa = 7.0HH279 pKa = 5.94NFGGMIHH286 pKa = 6.93GFFNMIADD294 pKa = 4.2PVDD297 pKa = 2.91IDD299 pKa = 4.18AAHH302 pKa = 6.64EE303 pKa = 4.4AYY305 pKa = 10.38DD306 pKa = 4.51AVVADD311 pKa = 5.05LDD313 pKa = 4.89GMLDD317 pKa = 3.65DD318 pKa = 5.06
MM1 pKa = 6.48TAEE4 pKa = 4.91RR5 pKa = 11.84SPEE8 pKa = 3.93PDD10 pKa = 3.22PQVEE14 pKa = 4.24QFLQLYY20 pKa = 10.62DD21 pKa = 3.89SMDD24 pKa = 3.44VPEE27 pKa = 4.63LHH29 pKa = 6.74EE30 pKa = 4.69LSPPEE35 pKa = 3.78ARR37 pKa = 11.84EE38 pKa = 3.86VQEE41 pKa = 4.53QLSQADD47 pKa = 3.94PDD49 pKa = 3.62IDD51 pKa = 4.08LPTVEE56 pKa = 5.73DD57 pKa = 3.83RR58 pKa = 11.84TIDD61 pKa = 3.94GPDD64 pKa = 3.18GEE66 pKa = 4.56IPIRR70 pKa = 11.84LYY72 pKa = 10.83DD73 pKa = 3.69PRR75 pKa = 11.84TEE77 pKa = 4.08EE78 pKa = 4.21TEE80 pKa = 4.04TPLILFYY87 pKa = 10.87HH88 pKa = 6.6GGGFVVGSLDD98 pKa = 3.58THH100 pKa = 7.33DD101 pKa = 4.41GPCRR105 pKa = 11.84KK106 pKa = 9.21LAAEE110 pKa = 3.91TGYY113 pKa = 10.07PVVSVDD119 pKa = 3.24YY120 pKa = 10.49RR121 pKa = 11.84LAPEE125 pKa = 4.58HH126 pKa = 6.76PFPAGLNDD134 pKa = 4.41CYY136 pKa = 11.2AALEE140 pKa = 4.37WASEE144 pKa = 3.95TDD146 pKa = 3.19EE147 pKa = 5.37LGIDD151 pKa = 3.37SGQIVVAGDD160 pKa = 3.48SAGGNFAAAVSLLSRR175 pKa = 11.84DD176 pKa = 3.22RR177 pKa = 11.84DD178 pKa = 3.85GPEE181 pKa = 3.12IAAQVLVYY189 pKa = 10.07PVTGDD194 pKa = 3.31AAEE197 pKa = 4.24TDD199 pKa = 4.17SYY201 pKa = 11.21DD202 pKa = 3.82QNGEE206 pKa = 4.25GYY208 pKa = 10.43FLTTEE213 pKa = 4.01TMEE216 pKa = 4.3WFHH219 pKa = 6.98DD220 pKa = 3.84QYY222 pKa = 11.51LDD224 pKa = 4.52DD225 pKa = 6.43PIDD228 pKa = 3.66EE229 pKa = 4.54GNIYY233 pKa = 9.7VAPRR237 pKa = 11.84LAADD241 pKa = 4.89LSDD244 pKa = 4.79LPPATVVTAGYY255 pKa = 10.56DD256 pKa = 3.49PLRR259 pKa = 11.84DD260 pKa = 3.96DD261 pKa = 3.97GALYY265 pKa = 10.82AEE267 pKa = 4.74RR268 pKa = 11.84LRR270 pKa = 11.84DD271 pKa = 3.48AGVEE275 pKa = 4.24VSHH278 pKa = 7.0HH279 pKa = 5.94NFGGMIHH286 pKa = 6.93GFFNMIADD294 pKa = 4.2PVDD297 pKa = 2.91IDD299 pKa = 4.18AAHH302 pKa = 6.64EE303 pKa = 4.4AYY305 pKa = 10.38DD306 pKa = 4.51AVVADD311 pKa = 5.05LDD313 pKa = 4.89GMLDD317 pKa = 3.65DD318 pKa = 5.06
Molecular weight: 34.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B0T7Z4|A0A6B0T7Z4_9EURY Uncharacterized protein OS=Halovenus sp. WSH3 OX=2692199 GN=GRX03_12020 PE=4 SV=1
MM1 pKa = 7.26TPRR4 pKa = 11.84SRR6 pKa = 11.84ALGVDD11 pKa = 4.57FGLGAGAVGLGALAGGTPQSLDD33 pKa = 3.0GFLIGGIGLFAFVATLYY50 pKa = 10.52VAEE53 pKa = 4.09QTKK56 pKa = 10.5VLVLIEE62 pKa = 4.1RR63 pKa = 11.84TRR65 pKa = 11.84PTSLVVACVTLAAVGVALVVGQVVVEE91 pKa = 4.85TPAATLLVGMGTGLIGYY108 pKa = 7.19RR109 pKa = 11.84LWFGLVAPVPPKK121 pKa = 10.63RR122 pKa = 11.84LDD124 pKa = 3.33QASVWGSRR132 pKa = 11.84RR133 pKa = 11.84NRR135 pKa = 11.84KK136 pKa = 8.63
MM1 pKa = 7.26TPRR4 pKa = 11.84SRR6 pKa = 11.84ALGVDD11 pKa = 4.57FGLGAGAVGLGALAGGTPQSLDD33 pKa = 3.0GFLIGGIGLFAFVATLYY50 pKa = 10.52VAEE53 pKa = 4.09QTKK56 pKa = 10.5VLVLIEE62 pKa = 4.1RR63 pKa = 11.84TRR65 pKa = 11.84PTSLVVACVTLAAVGVALVVGQVVVEE91 pKa = 4.85TPAATLLVGMGTGLIGYY108 pKa = 7.19RR109 pKa = 11.84LWFGLVAPVPPKK121 pKa = 10.63RR122 pKa = 11.84LDD124 pKa = 3.33QASVWGSRR132 pKa = 11.84RR133 pKa = 11.84NRR135 pKa = 11.84KK136 pKa = 8.63
Molecular weight: 13.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
956425 |
41 |
2990 |
300.3 |
32.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.178 ± 0.054 | 0.734 ± 0.013 |
8.085 ± 0.043 | 9.47 ± 0.064 |
3.232 ± 0.029 | 8.492 ± 0.045 |
1.905 ± 0.022 | 4.527 ± 0.032 |
1.808 ± 0.027 | 8.892 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.722 ± 0.019 | 2.37 ± 0.026 |
4.631 ± 0.028 | 2.977 ± 0.023 |
6.561 ± 0.046 | 5.657 ± 0.035 |
6.481 ± 0.04 | 8.512 ± 0.043 |
1.06 ± 0.018 | 2.707 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
