
Acinetobacter populi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
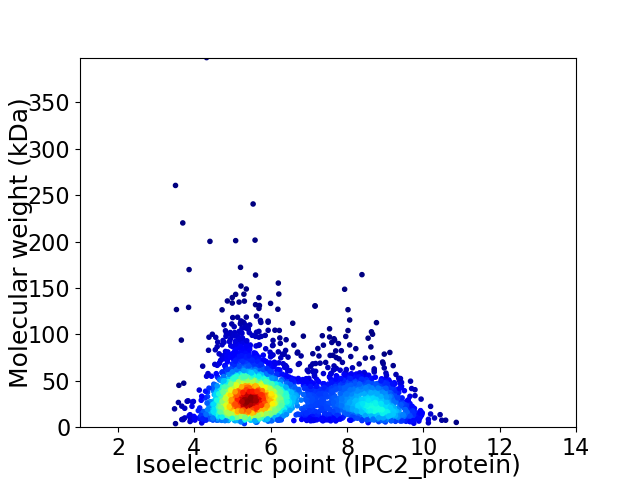
Virtual 2D-PAGE plot for 3291 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z9YYX0|A0A1Z9YYX0_9GAMM Uncharacterized protein OS=Acinetobacter populi OX=1582270 GN=CAP51_06270 PE=4 SV=1
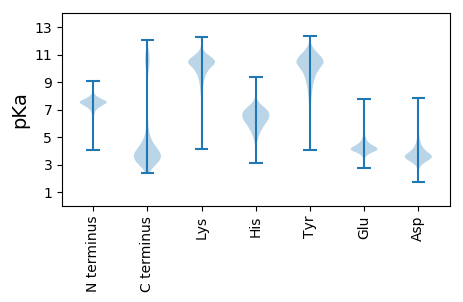
MM1 pKa = 7.84DD2 pKa = 4.11TTPIISGSTGFDD14 pKa = 2.99ILEE17 pKa = 4.27GEE19 pKa = 4.34YY20 pKa = 11.24LEE22 pKa = 4.26VTVNSKK28 pKa = 9.26TYY30 pKa = 10.38SSQTGAVVIDD40 pKa = 3.98FDD42 pKa = 4.39NNTWYY47 pKa = 10.77VQIPDD52 pKa = 4.03SDD54 pKa = 4.04ALPIGTYY61 pKa = 9.95NVSAVLKK68 pKa = 9.35NAEE71 pKa = 4.14GEE73 pKa = 4.2ITQDD77 pKa = 3.09DD78 pKa = 4.33TTNEE82 pKa = 4.18LVVSPSPTINFTATAATSDD101 pKa = 3.77DD102 pKa = 3.71TGTALTISEE111 pKa = 4.14NGTWRR116 pKa = 11.84ILSNSTVFTQNATDD130 pKa = 4.08PSTLGSFTSIAISGADD146 pKa = 3.43RR147 pKa = 11.84QQQSSFIDD155 pKa = 3.85FDD157 pKa = 4.65RR158 pKa = 11.84DD159 pKa = 3.11GLMDD163 pKa = 4.12ILGADD168 pKa = 3.3TSFANGQQSFKK179 pKa = 10.96YY180 pKa = 9.88NADD183 pKa = 2.79GTYY186 pKa = 9.78TVFQIGSFGISGQTNDD202 pKa = 4.22ANGNTYY208 pKa = 8.78VWYY211 pKa = 10.42GGVAGIDD218 pKa = 3.49INGDD222 pKa = 3.54GYY224 pKa = 11.75VDD226 pKa = 3.34IVYY229 pKa = 10.74GDD231 pKa = 3.79EE232 pKa = 4.35TPNDD236 pKa = 3.38AEE238 pKa = 4.35TRR240 pKa = 11.84GGYY243 pKa = 7.68DD244 pKa = 2.84TTFVLNTNGTILGFNKK260 pKa = 10.17SGAYY264 pKa = 10.1VYY266 pKa = 9.29TQTNQDD272 pKa = 2.74GVAATNSGNPTPDD285 pKa = 3.18RR286 pKa = 11.84EE287 pKa = 4.32IAGVDD292 pKa = 3.94LNNDD296 pKa = 3.48GYY298 pKa = 11.86VDD300 pKa = 3.69IVYY303 pKa = 10.31HH304 pKa = 5.67GTAGTNTTSTGGSSGVSTRR323 pKa = 11.84LVVVNNGVDD332 pKa = 3.51ANGNTTLTNTQIVTDD347 pKa = 4.19VFNGDD352 pKa = 3.26NGTTNVYY359 pKa = 9.97TSLTWADD366 pKa = 3.97FNGDD370 pKa = 3.01GYY372 pKa = 10.9MDD374 pKa = 4.62LFIGGLTGTGTAANSEE390 pKa = 4.24IYY392 pKa = 11.04YY393 pKa = 10.62NDD395 pKa = 3.44GTGKK399 pKa = 10.67LVTTTNGVGTGTNVQTLTDD418 pKa = 3.7ATNSNTSLAVDD429 pKa = 3.48WNGDD433 pKa = 3.3GKK435 pKa = 10.66IDD437 pKa = 3.81IIEE440 pKa = 4.13IAGISQGNSAANVSSADD457 pKa = 3.36NKK459 pKa = 10.8GILWLNGGTNASGQVNWTSEE479 pKa = 4.37TILAQANIRR488 pKa = 11.84PGAASTYY495 pKa = 9.49RR496 pKa = 11.84FVSGAQVVDD505 pKa = 4.2LDD507 pKa = 4.02YY508 pKa = 11.67DD509 pKa = 4.08GDD511 pKa = 3.62QDD513 pKa = 4.31LVVFRR518 pKa = 11.84SEE520 pKa = 4.23AGATSYY526 pKa = 10.94IEE528 pKa = 4.15NKK530 pKa = 10.38SVIQDD535 pKa = 3.4GTSIILNIRR544 pKa = 11.84DD545 pKa = 3.59KK546 pKa = 11.61NGINAYY552 pKa = 9.83YY553 pKa = 10.05GNTVALIDD561 pKa = 3.88EE562 pKa = 4.56ATGQVVATQIINAQSGVNTQNSTGLVYY589 pKa = 9.84FYY591 pKa = 11.28GLDD594 pKa = 3.35ASKK597 pKa = 10.75SYY599 pKa = 11.09SAVILSNGNDD609 pKa = 3.36YY610 pKa = 11.62GGISSITLGSSVNTIEE626 pKa = 4.93NVNEE630 pKa = 3.66TWAGLKK636 pKa = 10.24AVEE639 pKa = 4.42KK640 pKa = 9.59NHH642 pKa = 6.82AYY644 pKa = 10.75VLTTEE649 pKa = 4.29NGTAASSTATAATDD663 pKa = 3.44GTNTVGILGTGYY675 pKa = 11.0NDD677 pKa = 3.4TLYY680 pKa = 9.94ATAGTHH686 pKa = 5.74VYY688 pKa = 10.29NGAGGSEE695 pKa = 4.26LVSGVNTWSDD705 pKa = 3.26TGGMDD710 pKa = 3.62IVDD713 pKa = 3.79YY714 pKa = 10.95KK715 pKa = 11.13LAGSTAITVDD725 pKa = 3.97LSITTAQNTGFGTATFVNIEE745 pKa = 4.34GIAGSSSNDD754 pKa = 3.12TFTDD758 pKa = 3.31NAGNNQFEE766 pKa = 4.43GRR768 pKa = 11.84GGNDD772 pKa = 2.48IFNLINGGNDD782 pKa = 2.89TLLYY786 pKa = 10.35KK787 pKa = 10.79VLDD790 pKa = 4.0AANATGGNGSDD801 pKa = 3.6VVNGFHH807 pKa = 6.7VGTWEE812 pKa = 3.86ATPNADD818 pKa = 5.61RR819 pKa = 11.84IDD821 pKa = 4.0LSEE824 pKa = 4.37LLIGYY829 pKa = 6.79TGSVTPASYY838 pKa = 11.05INGTPTLAADD848 pKa = 3.62AEE850 pKa = 4.2IRR852 pKa = 11.84NYY854 pKa = 11.05LSVQSDD860 pKa = 3.75GTNTTISIDD869 pKa = 3.43RR870 pKa = 11.84DD871 pKa = 3.84GAGGAYY877 pKa = 10.3SSTTLVTLNDD887 pKa = 3.02VDD889 pKa = 3.98TTLEE893 pKa = 3.95KK894 pKa = 10.97LLANHH899 pKa = 6.45QIIIGG904 pKa = 3.69
MM1 pKa = 7.84DD2 pKa = 4.11TTPIISGSTGFDD14 pKa = 2.99ILEE17 pKa = 4.27GEE19 pKa = 4.34YY20 pKa = 11.24LEE22 pKa = 4.26VTVNSKK28 pKa = 9.26TYY30 pKa = 10.38SSQTGAVVIDD40 pKa = 3.98FDD42 pKa = 4.39NNTWYY47 pKa = 10.77VQIPDD52 pKa = 4.03SDD54 pKa = 4.04ALPIGTYY61 pKa = 9.95NVSAVLKK68 pKa = 9.35NAEE71 pKa = 4.14GEE73 pKa = 4.2ITQDD77 pKa = 3.09DD78 pKa = 4.33TTNEE82 pKa = 4.18LVVSPSPTINFTATAATSDD101 pKa = 3.77DD102 pKa = 3.71TGTALTISEE111 pKa = 4.14NGTWRR116 pKa = 11.84ILSNSTVFTQNATDD130 pKa = 4.08PSTLGSFTSIAISGADD146 pKa = 3.43RR147 pKa = 11.84QQQSSFIDD155 pKa = 3.85FDD157 pKa = 4.65RR158 pKa = 11.84DD159 pKa = 3.11GLMDD163 pKa = 4.12ILGADD168 pKa = 3.3TSFANGQQSFKK179 pKa = 10.96YY180 pKa = 9.88NADD183 pKa = 2.79GTYY186 pKa = 9.78TVFQIGSFGISGQTNDD202 pKa = 4.22ANGNTYY208 pKa = 8.78VWYY211 pKa = 10.42GGVAGIDD218 pKa = 3.49INGDD222 pKa = 3.54GYY224 pKa = 11.75VDD226 pKa = 3.34IVYY229 pKa = 10.74GDD231 pKa = 3.79EE232 pKa = 4.35TPNDD236 pKa = 3.38AEE238 pKa = 4.35TRR240 pKa = 11.84GGYY243 pKa = 7.68DD244 pKa = 2.84TTFVLNTNGTILGFNKK260 pKa = 10.17SGAYY264 pKa = 10.1VYY266 pKa = 9.29TQTNQDD272 pKa = 2.74GVAATNSGNPTPDD285 pKa = 3.18RR286 pKa = 11.84EE287 pKa = 4.32IAGVDD292 pKa = 3.94LNNDD296 pKa = 3.48GYY298 pKa = 11.86VDD300 pKa = 3.69IVYY303 pKa = 10.31HH304 pKa = 5.67GTAGTNTTSTGGSSGVSTRR323 pKa = 11.84LVVVNNGVDD332 pKa = 3.51ANGNTTLTNTQIVTDD347 pKa = 4.19VFNGDD352 pKa = 3.26NGTTNVYY359 pKa = 9.97TSLTWADD366 pKa = 3.97FNGDD370 pKa = 3.01GYY372 pKa = 10.9MDD374 pKa = 4.62LFIGGLTGTGTAANSEE390 pKa = 4.24IYY392 pKa = 11.04YY393 pKa = 10.62NDD395 pKa = 3.44GTGKK399 pKa = 10.67LVTTTNGVGTGTNVQTLTDD418 pKa = 3.7ATNSNTSLAVDD429 pKa = 3.48WNGDD433 pKa = 3.3GKK435 pKa = 10.66IDD437 pKa = 3.81IIEE440 pKa = 4.13IAGISQGNSAANVSSADD457 pKa = 3.36NKK459 pKa = 10.8GILWLNGGTNASGQVNWTSEE479 pKa = 4.37TILAQANIRR488 pKa = 11.84PGAASTYY495 pKa = 9.49RR496 pKa = 11.84FVSGAQVVDD505 pKa = 4.2LDD507 pKa = 4.02YY508 pKa = 11.67DD509 pKa = 4.08GDD511 pKa = 3.62QDD513 pKa = 4.31LVVFRR518 pKa = 11.84SEE520 pKa = 4.23AGATSYY526 pKa = 10.94IEE528 pKa = 4.15NKK530 pKa = 10.38SVIQDD535 pKa = 3.4GTSIILNIRR544 pKa = 11.84DD545 pKa = 3.59KK546 pKa = 11.61NGINAYY552 pKa = 9.83YY553 pKa = 10.05GNTVALIDD561 pKa = 3.88EE562 pKa = 4.56ATGQVVATQIINAQSGVNTQNSTGLVYY589 pKa = 9.84FYY591 pKa = 11.28GLDD594 pKa = 3.35ASKK597 pKa = 10.75SYY599 pKa = 11.09SAVILSNGNDD609 pKa = 3.36YY610 pKa = 11.62GGISSITLGSSVNTIEE626 pKa = 4.93NVNEE630 pKa = 3.66TWAGLKK636 pKa = 10.24AVEE639 pKa = 4.42KK640 pKa = 9.59NHH642 pKa = 6.82AYY644 pKa = 10.75VLTTEE649 pKa = 4.29NGTAASSTATAATDD663 pKa = 3.44GTNTVGILGTGYY675 pKa = 11.0NDD677 pKa = 3.4TLYY680 pKa = 9.94ATAGTHH686 pKa = 5.74VYY688 pKa = 10.29NGAGGSEE695 pKa = 4.26LVSGVNTWSDD705 pKa = 3.26TGGMDD710 pKa = 3.62IVDD713 pKa = 3.79YY714 pKa = 10.95KK715 pKa = 11.13LAGSTAITVDD725 pKa = 3.97LSITTAQNTGFGTATFVNIEE745 pKa = 4.34GIAGSSSNDD754 pKa = 3.12TFTDD758 pKa = 3.31NAGNNQFEE766 pKa = 4.43GRR768 pKa = 11.84GGNDD772 pKa = 2.48IFNLINGGNDD782 pKa = 2.89TLLYY786 pKa = 10.35KK787 pKa = 10.79VLDD790 pKa = 4.0AANATGGNGSDD801 pKa = 3.6VVNGFHH807 pKa = 6.7VGTWEE812 pKa = 3.86ATPNADD818 pKa = 5.61RR819 pKa = 11.84IDD821 pKa = 4.0LSEE824 pKa = 4.37LLIGYY829 pKa = 6.79TGSVTPASYY838 pKa = 11.05INGTPTLAADD848 pKa = 3.62AEE850 pKa = 4.2IRR852 pKa = 11.84NYY854 pKa = 11.05LSVQSDD860 pKa = 3.75GTNTTISIDD869 pKa = 3.43RR870 pKa = 11.84DD871 pKa = 3.84GAGGAYY877 pKa = 10.3SSTTLVTLNDD887 pKa = 3.02VDD889 pKa = 3.98TTLEE893 pKa = 3.95KK894 pKa = 10.97LLANHH899 pKa = 6.45QIIIGG904 pKa = 3.69
Molecular weight: 93.85 kDa
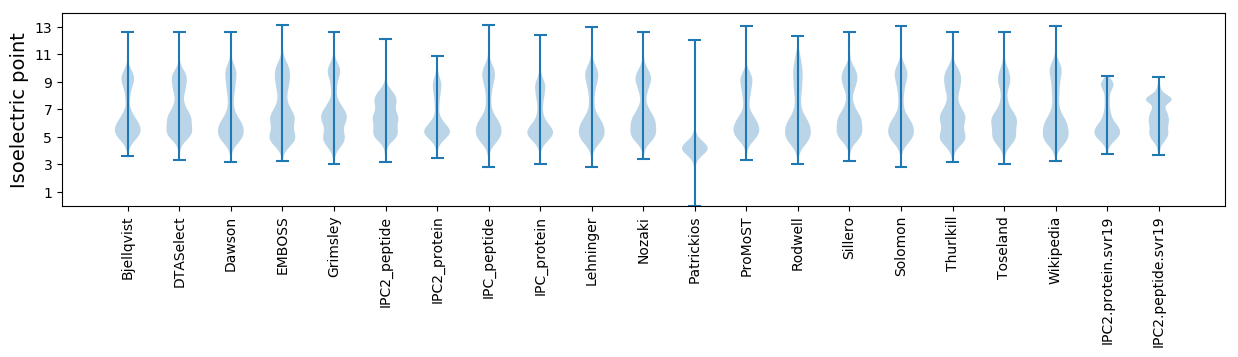
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z9YVZ0|A0A1Z9YVZ0_9GAMM GNAT family N-acetyltransferase OS=Acinetobacter populi OX=1582270 GN=CAP51_14005 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064362 |
33 |
3790 |
323.4 |
36.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.668 ± 0.047 | 0.924 ± 0.014 |
5.368 ± 0.031 | 5.514 ± 0.039 |
4.211 ± 0.035 | 6.574 ± 0.05 |
2.418 ± 0.032 | 7.145 ± 0.037 |
5.292 ± 0.036 | 10.442 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.024 | 4.505 ± 0.047 |
3.889 ± 0.029 | 5.952 ± 0.053 |
4.253 ± 0.034 | 6.096 ± 0.042 |
5.417 ± 0.048 | 6.379 ± 0.036 |
1.27 ± 0.018 | 3.387 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
