
Beihai sobemo-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
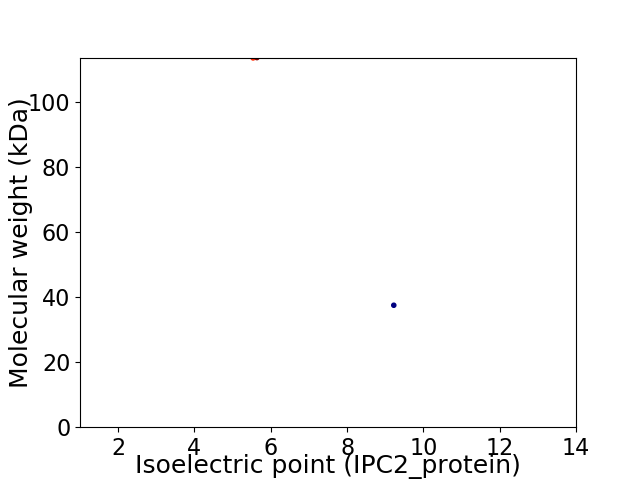
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEE8|A0A1L3KEE8_9VIRU Putative capsid protein OS=Beihai sobemo-like virus 8 OX=1922705 PE=4 SV=1
MM1 pKa = 7.75AFTVTISDD9 pKa = 3.89EE10 pKa = 4.64LIEE13 pKa = 4.14SAIWSFFGGGAMFLALKK30 pKa = 10.33IGVFGMLWEE39 pKa = 4.3AASWLLAWVWFVVQVVARR57 pKa = 11.84RR58 pKa = 11.84LAFQLQLGALPVAACKK74 pKa = 9.92TGFIIVGEE82 pKa = 4.01KK83 pKa = 10.07AYY85 pKa = 11.0YY86 pKa = 9.64RR87 pKa = 11.84DD88 pKa = 3.84PVDD91 pKa = 3.34LRR93 pKa = 11.84MYY95 pKa = 10.68SVVPHH100 pKa = 6.81TATCTDD106 pKa = 2.74GKK108 pKa = 10.15MVLKK112 pKa = 10.38DD113 pKa = 3.59YY114 pKa = 11.11RR115 pKa = 11.84LVPLGGDD122 pKa = 3.28AKK124 pKa = 10.56VAEE127 pKa = 4.4SALFGSDD134 pKa = 3.48PLTKK138 pKa = 10.11RR139 pKa = 11.84SSAPNDD145 pKa = 3.47YY146 pKa = 10.56VSLYY150 pKa = 10.08IEE152 pKa = 3.92QDD154 pKa = 3.12GEE156 pKa = 4.07YY157 pKa = 9.48HH158 pKa = 6.13YY159 pKa = 11.29LGGGYY164 pKa = 9.46RR165 pKa = 11.84EE166 pKa = 4.23GLFLITAAHH175 pKa = 7.06ALTEE179 pKa = 4.61SEE181 pKa = 4.19GHH183 pKa = 5.43TLALSRR189 pKa = 11.84DD190 pKa = 3.3GVKK193 pKa = 10.28FYY195 pKa = 11.17RR196 pKa = 11.84PAGVKK201 pKa = 9.11RR202 pKa = 11.84HH203 pKa = 5.16YY204 pKa = 10.76ARR206 pKa = 11.84EE207 pKa = 4.49GYY209 pKa = 9.86ARR211 pKa = 11.84CTGDD215 pKa = 3.77DD216 pKa = 3.2VGAFEE221 pKa = 6.05LSAADD226 pKa = 3.55WAASGVRR233 pKa = 11.84SVKK236 pKa = 9.91PSVYY240 pKa = 10.09SAVGQTRR247 pKa = 11.84VEE249 pKa = 3.93VFGRR253 pKa = 11.84SRR255 pKa = 11.84QGVLQAGVGALLPPTEE271 pKa = 4.02KK272 pKa = 10.18QKK274 pKa = 11.16RR275 pKa = 11.84LGIVPHH281 pKa = 6.15SASTIRR287 pKa = 11.84GFSGSPVYY295 pKa = 10.08STGEE299 pKa = 3.69SGRR302 pKa = 11.84RR303 pKa = 11.84IVGLHH308 pKa = 5.5IAGGCEE314 pKa = 4.01SDD316 pKa = 3.47NKK318 pKa = 10.95NYY320 pKa = 8.28MASVHH325 pKa = 6.51EE326 pKa = 4.13IHH328 pKa = 7.34FLRR331 pKa = 11.84QKK333 pKa = 10.66LGLVPEE339 pKa = 4.32PQLRR343 pKa = 11.84AEE345 pKa = 4.15VSPNTKK351 pKa = 8.75HH352 pKa = 5.7FQYY355 pKa = 10.5NRR357 pKa = 11.84HH358 pKa = 6.29DD359 pKa = 3.89RR360 pKa = 11.84DD361 pKa = 3.35SDD363 pKa = 3.71EE364 pKa = 5.21EE365 pKa = 4.08EE366 pKa = 5.56LDD368 pKa = 5.36DD369 pKa = 5.8EE370 pKa = 5.43LDD372 pKa = 3.59NANKK376 pKa = 10.34FGGTFGFYY384 pKa = 10.71DD385 pKa = 4.18SFAEE389 pKa = 4.16TAPVSAASAGAVTNPPPGLTPIVEE413 pKa = 4.49GAEE416 pKa = 4.24SEE418 pKa = 4.26PSPEE422 pKa = 3.83EE423 pKa = 3.89SGVGSEE429 pKa = 4.71APGAEE434 pKa = 4.07APSAEE439 pKa = 4.35GDD441 pKa = 3.46RR442 pKa = 11.84DD443 pKa = 4.07DD444 pKa = 6.73DD445 pKa = 3.96VLQVQDD451 pKa = 3.36AFEE454 pKa = 4.22EE455 pKa = 4.34EE456 pKa = 5.05EE457 pKa = 4.72YY458 pKa = 11.34VVLPDD463 pKa = 4.65IEE465 pKa = 5.19APPPPPAGAEE475 pKa = 3.86FVRR478 pKa = 11.84RR479 pKa = 11.84KK480 pKa = 9.97KK481 pKa = 10.51VGNIGRR487 pKa = 11.84FFGCTAFAGAAVAATNKK504 pKa = 9.45VFDD507 pKa = 4.71PAWMVCDD514 pKa = 4.51GPSLRR519 pKa = 11.84AAAGADD525 pKa = 3.51YY526 pKa = 11.23KK527 pKa = 11.86SMFEE531 pKa = 3.97STNFEE536 pKa = 4.25RR537 pKa = 11.84YY538 pKa = 8.99RR539 pKa = 11.84EE540 pKa = 4.04YY541 pKa = 11.22LSAARR546 pKa = 11.84AEE548 pKa = 4.19AAEE551 pKa = 4.67DD552 pKa = 3.37ARR554 pKa = 11.84TMKK557 pKa = 10.7GVDD560 pKa = 3.48GNDD563 pKa = 3.2VARR566 pKa = 11.84LFASGVSNRR575 pKa = 11.84SRR577 pKa = 11.84KK578 pKa = 9.3KK579 pKa = 9.34ALKK582 pKa = 9.82RR583 pKa = 11.84LPEE586 pKa = 4.36RR587 pKa = 11.84FLEE590 pKa = 4.86AIGRR594 pKa = 11.84LGLDD598 pKa = 3.47TKK600 pKa = 11.19AYY602 pKa = 8.51TGWANPPTGVEE613 pKa = 3.92AMEE616 pKa = 4.18QSFKK620 pKa = 10.97VQLGGVKK627 pKa = 8.09PTCWPEE633 pKa = 3.52ATRR636 pKa = 11.84KK637 pKa = 9.75QFVAKK642 pKa = 10.21EE643 pKa = 4.14GPLYY647 pKa = 10.69EE648 pKa = 5.5DD649 pKa = 4.85FLTEE653 pKa = 3.99VAKK656 pKa = 11.18YY657 pKa = 8.35PANRR661 pKa = 11.84YY662 pKa = 8.39NAFTGVHH669 pKa = 5.57ARR671 pKa = 11.84LTRR674 pKa = 11.84FVLGLDD680 pKa = 3.78GDD682 pKa = 4.54KK683 pKa = 11.1SAGWSQHH690 pKa = 4.84FRR692 pKa = 11.84PGTKK696 pKa = 9.79RR697 pKa = 11.84SWQDD701 pKa = 3.11KK702 pKa = 10.31EE703 pKa = 4.18GLEE706 pKa = 4.08LASYY710 pKa = 7.63LTRR713 pKa = 11.84CRR715 pKa = 11.84LLLRR719 pKa = 11.84AAVGPDD725 pKa = 2.96TMAQMTPAQLVGAGLSDD742 pKa = 3.73PRR744 pKa = 11.84TMFVKK749 pKa = 10.64VEE751 pKa = 3.83PHH753 pKa = 6.26NDD755 pKa = 2.88AKK757 pKa = 10.88VAAGRR762 pKa = 11.84WRR764 pKa = 11.84LIWGASLVDD773 pKa = 3.43VCTASVTCRR782 pKa = 11.84KK783 pKa = 9.01QDD785 pKa = 3.32KK786 pKa = 10.86LDD788 pKa = 3.38IEE790 pKa = 4.74QYY792 pKa = 10.13QGGPIPGGHH801 pKa = 5.0QQASGLGHH809 pKa = 7.21HH810 pKa = 7.29DD811 pKa = 3.66LGIDD815 pKa = 3.34RR816 pKa = 11.84LSRR819 pKa = 11.84EE820 pKa = 3.95FDD822 pKa = 3.27RR823 pKa = 11.84LLEE826 pKa = 4.38TGLEE830 pKa = 4.2VFDD833 pKa = 6.08ADD835 pKa = 3.7AQHH838 pKa = 6.71WDD840 pKa = 3.31MLVNRR845 pKa = 11.84DD846 pKa = 4.28SLYY849 pKa = 9.92ADD851 pKa = 3.5AWRR854 pKa = 11.84RR855 pKa = 11.84IILYY859 pKa = 10.27DD860 pKa = 3.79GSNKK864 pKa = 10.28DD865 pKa = 3.54VFEE868 pKa = 4.07MMALCEE874 pKa = 3.83AAANSAHH881 pKa = 5.42VVLVGGNLWEE891 pKa = 4.05ILKK894 pKa = 10.36PGITASGILSTTAQNSFIRR913 pKa = 11.84ALLYY917 pKa = 10.33SFVGIKK923 pKa = 9.98HH924 pKa = 5.89CVVAGDD930 pKa = 4.04DD931 pKa = 3.87AVGARR936 pKa = 11.84QKK938 pKa = 11.18GYY940 pKa = 8.46DD941 pKa = 3.58HH942 pKa = 7.34RR943 pKa = 11.84SALAEE948 pKa = 3.79YY949 pKa = 10.54GPIEE953 pKa = 4.19KK954 pKa = 10.12AVNVYY959 pKa = 9.13TPEE962 pKa = 5.19AGIEE966 pKa = 4.2FTSHH970 pKa = 6.95RR971 pKa = 11.84FTKK974 pKa = 10.55SAGVWTARR982 pKa = 11.84FLNLGKK988 pKa = 10.52ACARR992 pKa = 11.84LALGEE997 pKa = 4.2KK998 pKa = 9.36EE999 pKa = 4.21VRR1001 pKa = 11.84QDD1003 pKa = 3.45QLSGLLFCVRR1013 pKa = 11.84HH1014 pKa = 6.78DD1015 pKa = 3.51EE1016 pKa = 4.24AQIRR1020 pKa = 11.84NLGQVAEE1027 pKa = 4.1QMGWPIAGAEE1037 pKa = 4.13PVFLPCLDD1045 pKa = 3.59
MM1 pKa = 7.75AFTVTISDD9 pKa = 3.89EE10 pKa = 4.64LIEE13 pKa = 4.14SAIWSFFGGGAMFLALKK30 pKa = 10.33IGVFGMLWEE39 pKa = 4.3AASWLLAWVWFVVQVVARR57 pKa = 11.84RR58 pKa = 11.84LAFQLQLGALPVAACKK74 pKa = 9.92TGFIIVGEE82 pKa = 4.01KK83 pKa = 10.07AYY85 pKa = 11.0YY86 pKa = 9.64RR87 pKa = 11.84DD88 pKa = 3.84PVDD91 pKa = 3.34LRR93 pKa = 11.84MYY95 pKa = 10.68SVVPHH100 pKa = 6.81TATCTDD106 pKa = 2.74GKK108 pKa = 10.15MVLKK112 pKa = 10.38DD113 pKa = 3.59YY114 pKa = 11.11RR115 pKa = 11.84LVPLGGDD122 pKa = 3.28AKK124 pKa = 10.56VAEE127 pKa = 4.4SALFGSDD134 pKa = 3.48PLTKK138 pKa = 10.11RR139 pKa = 11.84SSAPNDD145 pKa = 3.47YY146 pKa = 10.56VSLYY150 pKa = 10.08IEE152 pKa = 3.92QDD154 pKa = 3.12GEE156 pKa = 4.07YY157 pKa = 9.48HH158 pKa = 6.13YY159 pKa = 11.29LGGGYY164 pKa = 9.46RR165 pKa = 11.84EE166 pKa = 4.23GLFLITAAHH175 pKa = 7.06ALTEE179 pKa = 4.61SEE181 pKa = 4.19GHH183 pKa = 5.43TLALSRR189 pKa = 11.84DD190 pKa = 3.3GVKK193 pKa = 10.28FYY195 pKa = 11.17RR196 pKa = 11.84PAGVKK201 pKa = 9.11RR202 pKa = 11.84HH203 pKa = 5.16YY204 pKa = 10.76ARR206 pKa = 11.84EE207 pKa = 4.49GYY209 pKa = 9.86ARR211 pKa = 11.84CTGDD215 pKa = 3.77DD216 pKa = 3.2VGAFEE221 pKa = 6.05LSAADD226 pKa = 3.55WAASGVRR233 pKa = 11.84SVKK236 pKa = 9.91PSVYY240 pKa = 10.09SAVGQTRR247 pKa = 11.84VEE249 pKa = 3.93VFGRR253 pKa = 11.84SRR255 pKa = 11.84QGVLQAGVGALLPPTEE271 pKa = 4.02KK272 pKa = 10.18QKK274 pKa = 11.16RR275 pKa = 11.84LGIVPHH281 pKa = 6.15SASTIRR287 pKa = 11.84GFSGSPVYY295 pKa = 10.08STGEE299 pKa = 3.69SGRR302 pKa = 11.84RR303 pKa = 11.84IVGLHH308 pKa = 5.5IAGGCEE314 pKa = 4.01SDD316 pKa = 3.47NKK318 pKa = 10.95NYY320 pKa = 8.28MASVHH325 pKa = 6.51EE326 pKa = 4.13IHH328 pKa = 7.34FLRR331 pKa = 11.84QKK333 pKa = 10.66LGLVPEE339 pKa = 4.32PQLRR343 pKa = 11.84AEE345 pKa = 4.15VSPNTKK351 pKa = 8.75HH352 pKa = 5.7FQYY355 pKa = 10.5NRR357 pKa = 11.84HH358 pKa = 6.29DD359 pKa = 3.89RR360 pKa = 11.84DD361 pKa = 3.35SDD363 pKa = 3.71EE364 pKa = 5.21EE365 pKa = 4.08EE366 pKa = 5.56LDD368 pKa = 5.36DD369 pKa = 5.8EE370 pKa = 5.43LDD372 pKa = 3.59NANKK376 pKa = 10.34FGGTFGFYY384 pKa = 10.71DD385 pKa = 4.18SFAEE389 pKa = 4.16TAPVSAASAGAVTNPPPGLTPIVEE413 pKa = 4.49GAEE416 pKa = 4.24SEE418 pKa = 4.26PSPEE422 pKa = 3.83EE423 pKa = 3.89SGVGSEE429 pKa = 4.71APGAEE434 pKa = 4.07APSAEE439 pKa = 4.35GDD441 pKa = 3.46RR442 pKa = 11.84DD443 pKa = 4.07DD444 pKa = 6.73DD445 pKa = 3.96VLQVQDD451 pKa = 3.36AFEE454 pKa = 4.22EE455 pKa = 4.34EE456 pKa = 5.05EE457 pKa = 4.72YY458 pKa = 11.34VVLPDD463 pKa = 4.65IEE465 pKa = 5.19APPPPPAGAEE475 pKa = 3.86FVRR478 pKa = 11.84RR479 pKa = 11.84KK480 pKa = 9.97KK481 pKa = 10.51VGNIGRR487 pKa = 11.84FFGCTAFAGAAVAATNKK504 pKa = 9.45VFDD507 pKa = 4.71PAWMVCDD514 pKa = 4.51GPSLRR519 pKa = 11.84AAAGADD525 pKa = 3.51YY526 pKa = 11.23KK527 pKa = 11.86SMFEE531 pKa = 3.97STNFEE536 pKa = 4.25RR537 pKa = 11.84YY538 pKa = 8.99RR539 pKa = 11.84EE540 pKa = 4.04YY541 pKa = 11.22LSAARR546 pKa = 11.84AEE548 pKa = 4.19AAEE551 pKa = 4.67DD552 pKa = 3.37ARR554 pKa = 11.84TMKK557 pKa = 10.7GVDD560 pKa = 3.48GNDD563 pKa = 3.2VARR566 pKa = 11.84LFASGVSNRR575 pKa = 11.84SRR577 pKa = 11.84KK578 pKa = 9.3KK579 pKa = 9.34ALKK582 pKa = 9.82RR583 pKa = 11.84LPEE586 pKa = 4.36RR587 pKa = 11.84FLEE590 pKa = 4.86AIGRR594 pKa = 11.84LGLDD598 pKa = 3.47TKK600 pKa = 11.19AYY602 pKa = 8.51TGWANPPTGVEE613 pKa = 3.92AMEE616 pKa = 4.18QSFKK620 pKa = 10.97VQLGGVKK627 pKa = 8.09PTCWPEE633 pKa = 3.52ATRR636 pKa = 11.84KK637 pKa = 9.75QFVAKK642 pKa = 10.21EE643 pKa = 4.14GPLYY647 pKa = 10.69EE648 pKa = 5.5DD649 pKa = 4.85FLTEE653 pKa = 3.99VAKK656 pKa = 11.18YY657 pKa = 8.35PANRR661 pKa = 11.84YY662 pKa = 8.39NAFTGVHH669 pKa = 5.57ARR671 pKa = 11.84LTRR674 pKa = 11.84FVLGLDD680 pKa = 3.78GDD682 pKa = 4.54KK683 pKa = 11.1SAGWSQHH690 pKa = 4.84FRR692 pKa = 11.84PGTKK696 pKa = 9.79RR697 pKa = 11.84SWQDD701 pKa = 3.11KK702 pKa = 10.31EE703 pKa = 4.18GLEE706 pKa = 4.08LASYY710 pKa = 7.63LTRR713 pKa = 11.84CRR715 pKa = 11.84LLLRR719 pKa = 11.84AAVGPDD725 pKa = 2.96TMAQMTPAQLVGAGLSDD742 pKa = 3.73PRR744 pKa = 11.84TMFVKK749 pKa = 10.64VEE751 pKa = 3.83PHH753 pKa = 6.26NDD755 pKa = 2.88AKK757 pKa = 10.88VAAGRR762 pKa = 11.84WRR764 pKa = 11.84LIWGASLVDD773 pKa = 3.43VCTASVTCRR782 pKa = 11.84KK783 pKa = 9.01QDD785 pKa = 3.32KK786 pKa = 10.86LDD788 pKa = 3.38IEE790 pKa = 4.74QYY792 pKa = 10.13QGGPIPGGHH801 pKa = 5.0QQASGLGHH809 pKa = 7.21HH810 pKa = 7.29DD811 pKa = 3.66LGIDD815 pKa = 3.34RR816 pKa = 11.84LSRR819 pKa = 11.84EE820 pKa = 3.95FDD822 pKa = 3.27RR823 pKa = 11.84LLEE826 pKa = 4.38TGLEE830 pKa = 4.2VFDD833 pKa = 6.08ADD835 pKa = 3.7AQHH838 pKa = 6.71WDD840 pKa = 3.31MLVNRR845 pKa = 11.84DD846 pKa = 4.28SLYY849 pKa = 9.92ADD851 pKa = 3.5AWRR854 pKa = 11.84RR855 pKa = 11.84IILYY859 pKa = 10.27DD860 pKa = 3.79GSNKK864 pKa = 10.28DD865 pKa = 3.54VFEE868 pKa = 4.07MMALCEE874 pKa = 3.83AAANSAHH881 pKa = 5.42VVLVGGNLWEE891 pKa = 4.05ILKK894 pKa = 10.36PGITASGILSTTAQNSFIRR913 pKa = 11.84ALLYY917 pKa = 10.33SFVGIKK923 pKa = 9.98HH924 pKa = 5.89CVVAGDD930 pKa = 4.04DD931 pKa = 3.87AVGARR936 pKa = 11.84QKK938 pKa = 11.18GYY940 pKa = 8.46DD941 pKa = 3.58HH942 pKa = 7.34RR943 pKa = 11.84SALAEE948 pKa = 3.79YY949 pKa = 10.54GPIEE953 pKa = 4.19KK954 pKa = 10.12AVNVYY959 pKa = 9.13TPEE962 pKa = 5.19AGIEE966 pKa = 4.2FTSHH970 pKa = 6.95RR971 pKa = 11.84FTKK974 pKa = 10.55SAGVWTARR982 pKa = 11.84FLNLGKK988 pKa = 10.52ACARR992 pKa = 11.84LALGEE997 pKa = 4.2KK998 pKa = 9.36EE999 pKa = 4.21VRR1001 pKa = 11.84QDD1003 pKa = 3.45QLSGLLFCVRR1013 pKa = 11.84HH1014 pKa = 6.78DD1015 pKa = 3.51EE1016 pKa = 4.24AQIRR1020 pKa = 11.84NLGQVAEE1027 pKa = 4.1QMGWPIAGAEE1037 pKa = 4.13PVFLPCLDD1045 pKa = 3.59
Molecular weight: 113.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEE8|A0A1L3KEE8_9VIRU Putative capsid protein OS=Beihai sobemo-like virus 8 OX=1922705 PE=4 SV=1
MM1 pKa = 7.43ARR3 pKa = 11.84NGRR6 pKa = 11.84RR7 pKa = 11.84PDD9 pKa = 3.24RR10 pKa = 11.84PPRR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84PPFRR21 pKa = 11.84RR22 pKa = 11.84GGRR25 pKa = 11.84RR26 pKa = 11.84STAEE30 pKa = 3.52RR31 pKa = 11.84VQAQGVGCVAPVPFGGKK48 pKa = 8.74RR49 pKa = 11.84VASIQGWNAFSPAHH63 pKa = 6.19LPLPRR68 pKa = 11.84SVGPYY73 pKa = 9.69CIVRR77 pKa = 11.84TTTRR81 pKa = 11.84VFSNKK86 pKa = 9.36KK87 pKa = 9.67FVTITCTLSEE97 pKa = 4.86DD98 pKa = 4.11GTWQAVCGFAEE109 pKa = 4.74GDD111 pKa = 3.77LGFGSAIDD119 pKa = 4.05GANNTQVLGSTFPGLTAGAGTSAITVVPSALSVQILNGEE158 pKa = 4.22ALQTTNGILAAGVVKK173 pKa = 9.06TQLALQGRR181 pKa = 11.84TEE183 pKa = 3.73TWEE186 pKa = 3.93AFSNRR191 pKa = 11.84FLAYY195 pKa = 7.33MAPRR199 pKa = 11.84LMSAGKK205 pKa = 9.58LALKK209 pKa = 9.48GVQMNSYY216 pKa = 8.76PLNMNVLSYY225 pKa = 8.71FAPLDD230 pKa = 3.5PTVFSGNITWKK241 pKa = 10.42SGEE244 pKa = 3.97NFQIYY249 pKa = 9.44PEE251 pKa = 4.51GWAPMCILNQDD262 pKa = 3.69AVDD265 pKa = 3.47ITYY268 pKa = 10.72LIAQEE273 pKa = 3.63WRR275 pKa = 11.84VRR277 pKa = 11.84FDD279 pKa = 3.72LANPAASAHH288 pKa = 5.47RR289 pKa = 11.84HH290 pKa = 5.68HH291 pKa = 7.04GVTSDD296 pKa = 3.74GLWDD300 pKa = 4.02KK301 pKa = 9.77MVSMASALGHH311 pKa = 6.08GVQDD315 pKa = 3.74LPEE318 pKa = 5.1FIANVGSAARR328 pKa = 11.84AANQTLRR335 pKa = 11.84MINAGTHH342 pKa = 5.25YY343 pKa = 10.98LPMLTGG349 pKa = 3.68
MM1 pKa = 7.43ARR3 pKa = 11.84NGRR6 pKa = 11.84RR7 pKa = 11.84PDD9 pKa = 3.24RR10 pKa = 11.84PPRR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84PPFRR21 pKa = 11.84RR22 pKa = 11.84GGRR25 pKa = 11.84RR26 pKa = 11.84STAEE30 pKa = 3.52RR31 pKa = 11.84VQAQGVGCVAPVPFGGKK48 pKa = 8.74RR49 pKa = 11.84VASIQGWNAFSPAHH63 pKa = 6.19LPLPRR68 pKa = 11.84SVGPYY73 pKa = 9.69CIVRR77 pKa = 11.84TTTRR81 pKa = 11.84VFSNKK86 pKa = 9.36KK87 pKa = 9.67FVTITCTLSEE97 pKa = 4.86DD98 pKa = 4.11GTWQAVCGFAEE109 pKa = 4.74GDD111 pKa = 3.77LGFGSAIDD119 pKa = 4.05GANNTQVLGSTFPGLTAGAGTSAITVVPSALSVQILNGEE158 pKa = 4.22ALQTTNGILAAGVVKK173 pKa = 9.06TQLALQGRR181 pKa = 11.84TEE183 pKa = 3.73TWEE186 pKa = 3.93AFSNRR191 pKa = 11.84FLAYY195 pKa = 7.33MAPRR199 pKa = 11.84LMSAGKK205 pKa = 9.58LALKK209 pKa = 9.48GVQMNSYY216 pKa = 8.76PLNMNVLSYY225 pKa = 8.71FAPLDD230 pKa = 3.5PTVFSGNITWKK241 pKa = 10.42SGEE244 pKa = 3.97NFQIYY249 pKa = 9.44PEE251 pKa = 4.51GWAPMCILNQDD262 pKa = 3.69AVDD265 pKa = 3.47ITYY268 pKa = 10.72LIAQEE273 pKa = 3.63WRR275 pKa = 11.84VRR277 pKa = 11.84FDD279 pKa = 3.72LANPAASAHH288 pKa = 5.47RR289 pKa = 11.84HH290 pKa = 5.68HH291 pKa = 7.04GVTSDD296 pKa = 3.74GLWDD300 pKa = 4.02KK301 pKa = 9.77MVSMASALGHH311 pKa = 6.08GVQDD315 pKa = 3.74LPEE318 pKa = 5.1FIANVGSAARR328 pKa = 11.84AANQTLRR335 pKa = 11.84MINAGTHH342 pKa = 5.25YY343 pKa = 10.98LPMLTGG349 pKa = 3.68
Molecular weight: 37.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394 |
349 |
1045 |
697.0 |
75.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.765 ± 0.144 | 1.435 ± 0.001 |
5.093 ± 0.918 | 5.739 ± 1.359 |
4.448 ± 0.071 | 9.828 ± 0.095 |
2.08 ± 0.171 | 3.3 ± 0.337 |
3.802 ± 0.714 | 8.465 ± 0.209 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.937 ± 0.439 | 3.156 ± 1.082 |
5.308 ± 0.471 | 3.443 ± 0.404 |
6.6 ± 0.267 | 6.169 ± 0.064 |
5.165 ± 0.945 | 7.604 ± 0.208 |
1.793 ± 0.1 | 2.869 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
