
Flagellimonas maritima
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flagellimonas
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
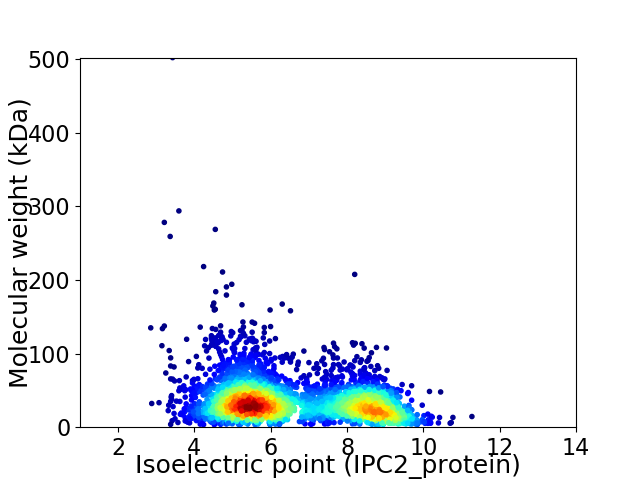
Virtual 2D-PAGE plot for 3376 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4LUC1|A0A2Z4LUC1_9FLAO Acyl-CoA 6-desaturase OS=Flagellimonas maritima OX=1383885 GN=HME9304_02346 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.23KK3 pKa = 10.32FYY5 pKa = 11.08ALFPILGLFFIACSDD20 pKa = 3.85DD21 pKa = 5.28DD22 pKa = 4.14EE23 pKa = 5.17TPSEE27 pKa = 4.39TTPTPDD33 pKa = 4.62PIVYY37 pKa = 9.36TSGSADD43 pKa = 3.32FSNYY47 pKa = 7.91VAVGNSLTAGFSDD60 pKa = 3.33NALFRR65 pKa = 11.84AGQEE69 pKa = 3.63ASFPNMLAANFALAGGGTFEE89 pKa = 4.9IPFMADD95 pKa = 2.77NLGGATINGQTDD107 pKa = 3.5ANMDD111 pKa = 3.77GAPDD115 pKa = 3.19IGNRR119 pKa = 11.84LILLFTPDD127 pKa = 4.09GPAPTPKK134 pKa = 9.68TGVGTTEE141 pKa = 3.81ITDD144 pKa = 3.81KK145 pKa = 11.37LSGPFNNMGVPGSKK159 pKa = 9.74SYY161 pKa = 11.06EE162 pKa = 3.98LLAPGYY168 pKa = 10.64GSLAGLAVGAANPYY182 pKa = 9.37FVRR185 pKa = 11.84FSSGEE190 pKa = 3.76AATVIGDD197 pKa = 4.28AAAQNPTFFSLWTGSNDD214 pKa = 2.65ILLYY218 pKa = 9.5ATSGGTGTFQSDD230 pKa = 3.93PMVSPADD237 pKa = 3.62YY238 pKa = 11.01ARR240 pKa = 11.84NDD242 pKa = 3.12ITNPTVFAGALDD254 pKa = 4.0AALQALTAGDD264 pKa = 3.88TDD266 pKa = 4.1GVIANLPNVTDD277 pKa = 3.53IPFFTTVPHH286 pKa = 6.28NPVPLDD292 pKa = 3.49EE293 pKa = 4.6ATAAQLNAAYY303 pKa = 10.31ALYY306 pKa = 10.83NGGLLQAQGFNLISEE321 pKa = 4.49EE322 pKa = 4.08EE323 pKa = 4.24VAKK326 pKa = 9.79RR327 pKa = 11.84TISFTAGEE335 pKa = 4.25GNAVVLLDD343 pKa = 4.76EE344 pKa = 5.72DD345 pKa = 4.17LTDD348 pKa = 3.73LTGVNPALINMRR360 pKa = 11.84QATEE364 pKa = 3.78EE365 pKa = 4.13DD366 pKa = 4.36LLVLTARR373 pKa = 11.84TFIGTEE379 pKa = 3.95AVVGDD384 pKa = 3.93PTTINGIAVPLADD397 pKa = 3.3QWVLTPEE404 pKa = 3.87EE405 pKa = 3.86QANIEE410 pKa = 4.34TALTAYY416 pKa = 9.87NQTITALATQYY427 pKa = 11.6DD428 pKa = 4.4LALVDD433 pKa = 4.23VNSLLSEE440 pKa = 3.95LRR442 pKa = 11.84EE443 pKa = 4.05NGIPLADD450 pKa = 4.01GSTVTATFATGGGFSLDD467 pKa = 3.48GVHH470 pKa = 7.01PSPRR474 pKa = 11.84GYY476 pKa = 10.95AIIANAFVAAINAKK490 pKa = 10.13YY491 pKa = 10.42GSNLPGVNPLDD502 pKa = 3.79YY503 pKa = 10.61TGLYY507 pKa = 9.52IDD509 pKa = 4.39
MM1 pKa = 7.48KK2 pKa = 10.23KK3 pKa = 10.32FYY5 pKa = 11.08ALFPILGLFFIACSDD20 pKa = 3.85DD21 pKa = 5.28DD22 pKa = 4.14EE23 pKa = 5.17TPSEE27 pKa = 4.39TTPTPDD33 pKa = 4.62PIVYY37 pKa = 9.36TSGSADD43 pKa = 3.32FSNYY47 pKa = 7.91VAVGNSLTAGFSDD60 pKa = 3.33NALFRR65 pKa = 11.84AGQEE69 pKa = 3.63ASFPNMLAANFALAGGGTFEE89 pKa = 4.9IPFMADD95 pKa = 2.77NLGGATINGQTDD107 pKa = 3.5ANMDD111 pKa = 3.77GAPDD115 pKa = 3.19IGNRR119 pKa = 11.84LILLFTPDD127 pKa = 4.09GPAPTPKK134 pKa = 9.68TGVGTTEE141 pKa = 3.81ITDD144 pKa = 3.81KK145 pKa = 11.37LSGPFNNMGVPGSKK159 pKa = 9.74SYY161 pKa = 11.06EE162 pKa = 3.98LLAPGYY168 pKa = 10.64GSLAGLAVGAANPYY182 pKa = 9.37FVRR185 pKa = 11.84FSSGEE190 pKa = 3.76AATVIGDD197 pKa = 4.28AAAQNPTFFSLWTGSNDD214 pKa = 2.65ILLYY218 pKa = 9.5ATSGGTGTFQSDD230 pKa = 3.93PMVSPADD237 pKa = 3.62YY238 pKa = 11.01ARR240 pKa = 11.84NDD242 pKa = 3.12ITNPTVFAGALDD254 pKa = 4.0AALQALTAGDD264 pKa = 3.88TDD266 pKa = 4.1GVIANLPNVTDD277 pKa = 3.53IPFFTTVPHH286 pKa = 6.28NPVPLDD292 pKa = 3.49EE293 pKa = 4.6ATAAQLNAAYY303 pKa = 10.31ALYY306 pKa = 10.83NGGLLQAQGFNLISEE321 pKa = 4.49EE322 pKa = 4.08EE323 pKa = 4.24VAKK326 pKa = 9.79RR327 pKa = 11.84TISFTAGEE335 pKa = 4.25GNAVVLLDD343 pKa = 4.76EE344 pKa = 5.72DD345 pKa = 4.17LTDD348 pKa = 3.73LTGVNPALINMRR360 pKa = 11.84QATEE364 pKa = 3.78EE365 pKa = 4.13DD366 pKa = 4.36LLVLTARR373 pKa = 11.84TFIGTEE379 pKa = 3.95AVVGDD384 pKa = 3.93PTTINGIAVPLADD397 pKa = 3.3QWVLTPEE404 pKa = 3.87EE405 pKa = 3.86QANIEE410 pKa = 4.34TALTAYY416 pKa = 9.87NQTITALATQYY427 pKa = 11.6DD428 pKa = 4.4LALVDD433 pKa = 4.23VNSLLSEE440 pKa = 3.95LRR442 pKa = 11.84EE443 pKa = 4.05NGIPLADD450 pKa = 4.01GSTVTATFATGGGFSLDD467 pKa = 3.48GVHH470 pKa = 7.01PSPRR474 pKa = 11.84GYY476 pKa = 10.95AIIANAFVAAINAKK490 pKa = 10.13YY491 pKa = 10.42GSNLPGVNPLDD502 pKa = 3.79YY503 pKa = 10.61TGLYY507 pKa = 9.52IDD509 pKa = 4.39
Molecular weight: 52.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4LSW2|A0A2Z4LSW2_9FLAO Uncharacterized protein OS=Flagellimonas maritima OX=1383885 GN=HME9304_01677 PE=4 SV=1
MM1 pKa = 8.08RR2 pKa = 11.84GQLQVASNNPKK13 pKa = 9.33FKK15 pKa = 10.75FFILHH20 pKa = 6.37PSPFIHH26 pKa = 6.62HH27 pKa = 7.13PSSITLHH34 pKa = 6.19PSPFIHH40 pKa = 6.61HH41 pKa = 7.03PSSVTHH47 pKa = 6.43HH48 pKa = 6.81PSPFTRR54 pKa = 11.84HH55 pKa = 5.94PSPIIRR61 pKa = 11.84HH62 pKa = 5.71PSPVTRR68 pKa = 11.84HH69 pKa = 5.8PSPVTRR75 pKa = 11.84HH76 pKa = 5.82PSPFTRR82 pKa = 11.84HH83 pKa = 5.94LSPFARR89 pKa = 11.84HH90 pKa = 6.24LSPIPRR96 pKa = 11.84HH97 pKa = 5.56PSPMILHH104 pKa = 6.74PSPFTRR110 pKa = 11.84HH111 pKa = 5.92RR112 pKa = 11.84SPFTRR117 pKa = 11.84HH118 pKa = 5.94PSPFTRR124 pKa = 11.84HH125 pKa = 5.12PP126 pKa = 3.52
MM1 pKa = 8.08RR2 pKa = 11.84GQLQVASNNPKK13 pKa = 9.33FKK15 pKa = 10.75FFILHH20 pKa = 6.37PSPFIHH26 pKa = 6.62HH27 pKa = 7.13PSSITLHH34 pKa = 6.19PSPFIHH40 pKa = 6.61HH41 pKa = 7.03PSSVTHH47 pKa = 6.43HH48 pKa = 6.81PSPFTRR54 pKa = 11.84HH55 pKa = 5.94PSPIIRR61 pKa = 11.84HH62 pKa = 5.71PSPVTRR68 pKa = 11.84HH69 pKa = 5.8PSPVTRR75 pKa = 11.84HH76 pKa = 5.82PSPFTRR82 pKa = 11.84HH83 pKa = 5.94LSPFARR89 pKa = 11.84HH90 pKa = 6.24LSPIPRR96 pKa = 11.84HH97 pKa = 5.56PSPMILHH104 pKa = 6.74PSPFTRR110 pKa = 11.84HH111 pKa = 5.92RR112 pKa = 11.84SPFTRR117 pKa = 11.84HH118 pKa = 5.94PSPFTRR124 pKa = 11.84HH125 pKa = 5.12PP126 pKa = 3.52
Molecular weight: 14.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1140652 |
29 |
4759 |
337.9 |
38.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.15 ± 0.037 | 0.733 ± 0.013 |
5.793 ± 0.048 | 6.713 ± 0.042 |
5.339 ± 0.032 | 6.729 ± 0.045 |
1.751 ± 0.021 | 7.786 ± 0.037 |
7.531 ± 0.069 | 9.436 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.219 ± 0.022 | 5.992 ± 0.036 |
3.509 ± 0.024 | 3.268 ± 0.02 |
3.709 ± 0.025 | 6.549 ± 0.034 |
5.596 ± 0.05 | 6.146 ± 0.031 |
1.096 ± 0.017 | 3.953 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
