
Badger associated gemykibivirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus badas1
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
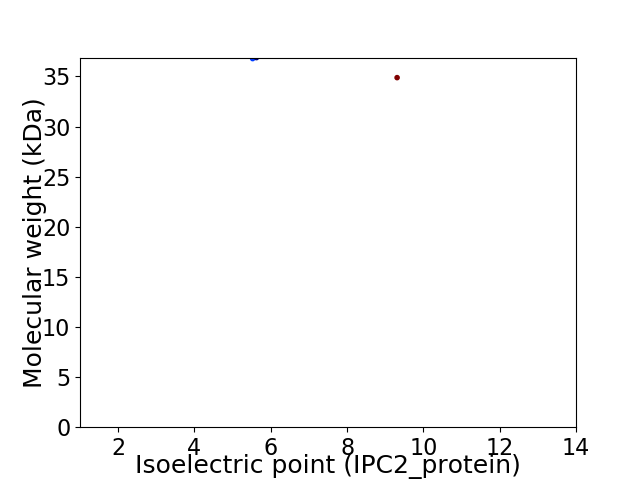
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3JHI0|A0A0E3JHI0_9VIRU Capsid OS=Badger associated gemykibivirus 1 OX=1634484 PE=4 SV=1
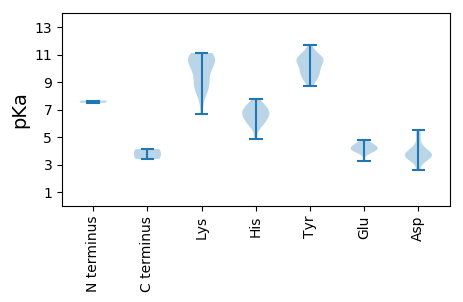
MM1 pKa = 7.49PAFYY5 pKa = 11.09VNSRR9 pKa = 11.84YY10 pKa = 10.25VLLTYY15 pKa = 10.05AQSGSLDD22 pKa = 3.52PFDD25 pKa = 5.26VVDD28 pKa = 4.55HH29 pKa = 6.55LASIHH34 pKa = 5.87GEE36 pKa = 4.06CIVGRR41 pKa = 11.84EE42 pKa = 3.96HH43 pKa = 7.09HH44 pKa = 7.01RR45 pKa = 11.84DD46 pKa = 3.52GGVHH50 pKa = 5.78LHH52 pKa = 6.47AFVHH56 pKa = 6.57FGRR59 pKa = 11.84KK60 pKa = 8.2FRR62 pKa = 11.84STSATVFDD70 pKa = 4.03VAGFHH75 pKa = 6.72PNISPSFGNPGRR87 pKa = 11.84GFDD90 pKa = 3.79YY91 pKa = 10.7AIKK94 pKa = 10.9DD95 pKa = 3.52GDD97 pKa = 3.5ICAGGLEE104 pKa = 4.21RR105 pKa = 11.84PGDD108 pKa = 3.78AGAPPTKK115 pKa = 10.17DD116 pKa = 2.73VWSQIVMAKK125 pKa = 10.43DD126 pKa = 3.44RR127 pKa = 11.84DD128 pKa = 4.24EE129 pKa = 4.36FWSLLRR135 pKa = 11.84QHH137 pKa = 6.17QPRR140 pKa = 11.84ALATSFVSLQKK151 pKa = 10.37YY152 pKa = 9.71ADD154 pKa = 2.82WHH156 pKa = 5.94FRR158 pKa = 11.84VDD160 pKa = 3.72PEE162 pKa = 4.77PYY164 pKa = 9.87QHH166 pKa = 7.18DD167 pKa = 4.08PSLGFYY173 pKa = 10.59LDD175 pKa = 5.14DD176 pKa = 4.17YY177 pKa = 10.79PEE179 pKa = 4.63LGSWVDD185 pKa = 3.91GYY187 pKa = 11.67LRR189 pKa = 11.84TNNTGMCAQSALSGLSGVGMSLVIYY214 pKa = 10.09GPSRR218 pKa = 11.84VGKK221 pKa = 6.67TVWARR226 pKa = 11.84SLGNHH231 pKa = 7.08AYY233 pKa = 10.43FGGLYY238 pKa = 10.49SLDD241 pKa = 3.48EE242 pKa = 4.37GVEE245 pKa = 4.12NVDD248 pKa = 3.31YY249 pKa = 11.29AVFDD253 pKa = 5.21DD254 pKa = 3.97IQGGLEE260 pKa = 3.85FFHH263 pKa = 7.45AYY265 pKa = 9.59KK266 pKa = 10.43FWLGHH271 pKa = 4.83QSQFYY276 pKa = 9.09ATDD279 pKa = 3.33KK280 pKa = 10.95YY281 pKa = 10.99KK282 pKa = 10.75SKK284 pKa = 11.11KK285 pKa = 8.89LIHH288 pKa = 6.74WGRR291 pKa = 11.84PAIWLANTDD300 pKa = 3.29PRR302 pKa = 11.84LEE304 pKa = 4.43KK305 pKa = 10.81GADD308 pKa = 3.83SDD310 pKa = 3.83WLEE313 pKa = 4.08ANCIFVCVTTPLYY326 pKa = 10.81SCQQ329 pKa = 3.41
MM1 pKa = 7.49PAFYY5 pKa = 11.09VNSRR9 pKa = 11.84YY10 pKa = 10.25VLLTYY15 pKa = 10.05AQSGSLDD22 pKa = 3.52PFDD25 pKa = 5.26VVDD28 pKa = 4.55HH29 pKa = 6.55LASIHH34 pKa = 5.87GEE36 pKa = 4.06CIVGRR41 pKa = 11.84EE42 pKa = 3.96HH43 pKa = 7.09HH44 pKa = 7.01RR45 pKa = 11.84DD46 pKa = 3.52GGVHH50 pKa = 5.78LHH52 pKa = 6.47AFVHH56 pKa = 6.57FGRR59 pKa = 11.84KK60 pKa = 8.2FRR62 pKa = 11.84STSATVFDD70 pKa = 4.03VAGFHH75 pKa = 6.72PNISPSFGNPGRR87 pKa = 11.84GFDD90 pKa = 3.79YY91 pKa = 10.7AIKK94 pKa = 10.9DD95 pKa = 3.52GDD97 pKa = 3.5ICAGGLEE104 pKa = 4.21RR105 pKa = 11.84PGDD108 pKa = 3.78AGAPPTKK115 pKa = 10.17DD116 pKa = 2.73VWSQIVMAKK125 pKa = 10.43DD126 pKa = 3.44RR127 pKa = 11.84DD128 pKa = 4.24EE129 pKa = 4.36FWSLLRR135 pKa = 11.84QHH137 pKa = 6.17QPRR140 pKa = 11.84ALATSFVSLQKK151 pKa = 10.37YY152 pKa = 9.71ADD154 pKa = 2.82WHH156 pKa = 5.94FRR158 pKa = 11.84VDD160 pKa = 3.72PEE162 pKa = 4.77PYY164 pKa = 9.87QHH166 pKa = 7.18DD167 pKa = 4.08PSLGFYY173 pKa = 10.59LDD175 pKa = 5.14DD176 pKa = 4.17YY177 pKa = 10.79PEE179 pKa = 4.63LGSWVDD185 pKa = 3.91GYY187 pKa = 11.67LRR189 pKa = 11.84TNNTGMCAQSALSGLSGVGMSLVIYY214 pKa = 10.09GPSRR218 pKa = 11.84VGKK221 pKa = 6.67TVWARR226 pKa = 11.84SLGNHH231 pKa = 7.08AYY233 pKa = 10.43FGGLYY238 pKa = 10.49SLDD241 pKa = 3.48EE242 pKa = 4.37GVEE245 pKa = 4.12NVDD248 pKa = 3.31YY249 pKa = 11.29AVFDD253 pKa = 5.21DD254 pKa = 3.97IQGGLEE260 pKa = 3.85FFHH263 pKa = 7.45AYY265 pKa = 9.59KK266 pKa = 10.43FWLGHH271 pKa = 4.83QSQFYY276 pKa = 9.09ATDD279 pKa = 3.33KK280 pKa = 10.95YY281 pKa = 10.99KK282 pKa = 10.75SKK284 pKa = 11.11KK285 pKa = 8.89LIHH288 pKa = 6.74WGRR291 pKa = 11.84PAIWLANTDD300 pKa = 3.29PRR302 pKa = 11.84LEE304 pKa = 4.43KK305 pKa = 10.81GADD308 pKa = 3.83SDD310 pKa = 3.83WLEE313 pKa = 4.08ANCIFVCVTTPLYY326 pKa = 10.81SCQQ329 pKa = 3.41
Molecular weight: 36.78 kDa
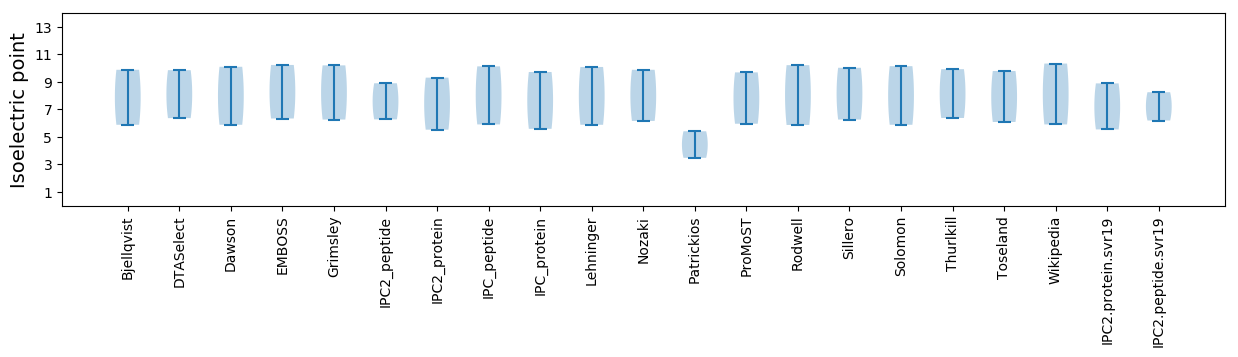
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3JHI0|A0A0E3JHI0_9VIRU Capsid OS=Badger associated gemykibivirus 1 OX=1634484 PE=4 SV=1
MM1 pKa = 7.65ALRR4 pKa = 11.84YY5 pKa = 9.44KK6 pKa = 10.58SRR8 pKa = 11.84SNYY11 pKa = 9.01RR12 pKa = 11.84RR13 pKa = 11.84IPRR16 pKa = 11.84SRR18 pKa = 11.84AKK20 pKa = 9.76YY21 pKa = 8.68RR22 pKa = 11.84ARR24 pKa = 11.84KK25 pKa = 9.17RR26 pKa = 11.84SAPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TYY34 pKa = 10.68RR35 pKa = 11.84KK36 pKa = 8.15RR37 pKa = 11.84QMSVRR42 pKa = 11.84RR43 pKa = 11.84IRR45 pKa = 11.84NITSRR50 pKa = 11.84KK51 pKa = 8.61CQDD54 pKa = 2.69NMMPITISDD63 pKa = 3.82DD64 pKa = 3.37GVTEE68 pKa = 4.16GTPGVGVTMVGDD80 pKa = 3.97FVYY83 pKa = 9.36TFCFVPSGRR92 pKa = 11.84MPGPQYY98 pKa = 10.58GASEE102 pKa = 4.18RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84ARR107 pKa = 11.84TFARR111 pKa = 11.84GYY113 pKa = 10.65KK114 pKa = 8.71EE115 pKa = 3.99TGLLEE120 pKa = 4.37TQTDD124 pKa = 3.97AQWIWRR130 pKa = 11.84RR131 pKa = 11.84IVFRR135 pKa = 11.84MRR137 pKa = 11.84LGKK140 pKa = 9.55WIGGFDD146 pKa = 3.64TLLDD150 pKa = 3.77PTQTLILDD158 pKa = 3.68HH159 pKa = 6.26TTNGMMRR166 pKa = 11.84TLWNLGNTTDD176 pKa = 5.11LNAIDD181 pKa = 5.53RR182 pKa = 11.84NDD184 pKa = 3.93ALDD187 pKa = 4.44KK188 pKa = 10.95ILFKK192 pKa = 10.43GTQGQDD198 pKa = 2.57WADD201 pKa = 3.52KK202 pKa = 8.92FTAPVDD208 pKa = 3.63PTSVYY213 pKa = 10.76LIRR216 pKa = 11.84DD217 pKa = 3.34KK218 pKa = 10.91TRR220 pKa = 11.84YY221 pKa = 9.44IGGSMAASGRR231 pKa = 11.84FHH233 pKa = 7.73RR234 pKa = 11.84VRR236 pKa = 11.84DD237 pKa = 3.88WYY239 pKa = 10.01PVNRR243 pKa = 11.84TLVYY247 pKa = 10.53EE248 pKa = 4.32DD249 pKa = 4.69TEE251 pKa = 4.57HH252 pKa = 6.55GQPPDD257 pKa = 3.31TATPWSDD264 pKa = 3.63EE265 pKa = 3.87SRR267 pKa = 11.84TTCGDD272 pKa = 3.54VVVLDD277 pKa = 3.93MFRR280 pKa = 11.84CASGDD285 pKa = 3.49VTDD288 pKa = 4.37TMSFSPEE295 pKa = 3.23GCYY298 pKa = 9.88YY299 pKa = 9.39WHH301 pKa = 7.15EE302 pKa = 4.13
MM1 pKa = 7.65ALRR4 pKa = 11.84YY5 pKa = 9.44KK6 pKa = 10.58SRR8 pKa = 11.84SNYY11 pKa = 9.01RR12 pKa = 11.84RR13 pKa = 11.84IPRR16 pKa = 11.84SRR18 pKa = 11.84AKK20 pKa = 9.76YY21 pKa = 8.68RR22 pKa = 11.84ARR24 pKa = 11.84KK25 pKa = 9.17RR26 pKa = 11.84SAPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TYY34 pKa = 10.68RR35 pKa = 11.84KK36 pKa = 8.15RR37 pKa = 11.84QMSVRR42 pKa = 11.84RR43 pKa = 11.84IRR45 pKa = 11.84NITSRR50 pKa = 11.84KK51 pKa = 8.61CQDD54 pKa = 2.69NMMPITISDD63 pKa = 3.82DD64 pKa = 3.37GVTEE68 pKa = 4.16GTPGVGVTMVGDD80 pKa = 3.97FVYY83 pKa = 9.36TFCFVPSGRR92 pKa = 11.84MPGPQYY98 pKa = 10.58GASEE102 pKa = 4.18RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84ARR107 pKa = 11.84TFARR111 pKa = 11.84GYY113 pKa = 10.65KK114 pKa = 8.71EE115 pKa = 3.99TGLLEE120 pKa = 4.37TQTDD124 pKa = 3.97AQWIWRR130 pKa = 11.84RR131 pKa = 11.84IVFRR135 pKa = 11.84MRR137 pKa = 11.84LGKK140 pKa = 9.55WIGGFDD146 pKa = 3.64TLLDD150 pKa = 3.77PTQTLILDD158 pKa = 3.68HH159 pKa = 6.26TTNGMMRR166 pKa = 11.84TLWNLGNTTDD176 pKa = 5.11LNAIDD181 pKa = 5.53RR182 pKa = 11.84NDD184 pKa = 3.93ALDD187 pKa = 4.44KK188 pKa = 10.95ILFKK192 pKa = 10.43GTQGQDD198 pKa = 2.57WADD201 pKa = 3.52KK202 pKa = 8.92FTAPVDD208 pKa = 3.63PTSVYY213 pKa = 10.76LIRR216 pKa = 11.84DD217 pKa = 3.34KK218 pKa = 10.91TRR220 pKa = 11.84YY221 pKa = 9.44IGGSMAASGRR231 pKa = 11.84FHH233 pKa = 7.73RR234 pKa = 11.84VRR236 pKa = 11.84DD237 pKa = 3.88WYY239 pKa = 10.01PVNRR243 pKa = 11.84TLVYY247 pKa = 10.53EE248 pKa = 4.32DD249 pKa = 4.69TEE251 pKa = 4.57HH252 pKa = 6.55GQPPDD257 pKa = 3.31TATPWSDD264 pKa = 3.63EE265 pKa = 3.87SRR267 pKa = 11.84TTCGDD272 pKa = 3.54VVVLDD277 pKa = 3.93MFRR280 pKa = 11.84CASGDD285 pKa = 3.49VTDD288 pKa = 4.37TMSFSPEE295 pKa = 3.23GCYY298 pKa = 9.88YY299 pKa = 9.39WHH301 pKa = 7.15EE302 pKa = 4.13
Molecular weight: 34.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631 |
302 |
329 |
315.5 |
35.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.498 ± 0.814 | 1.743 ± 0.059 |
7.924 ± 0.016 | 3.17 ± 0.129 |
4.913 ± 0.862 | 9.033 ± 0.737 |
3.011 ± 1.145 | 3.803 ± 0.34 |
3.645 ± 0.002 | 6.815 ± 1.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.976 | 2.853 ± 0.087 |
5.071 ± 0.071 | 3.17 ± 0.129 |
8.558 ± 2.732 | 6.656 ± 0.697 |
6.815 ± 2.342 | 6.339 ± 0.482 |
2.694 ± 0.031 | 4.754 ± 0.305 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
