
Flavonifractor sp. An10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Flavonifractor; unclassified Flavonifractor
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
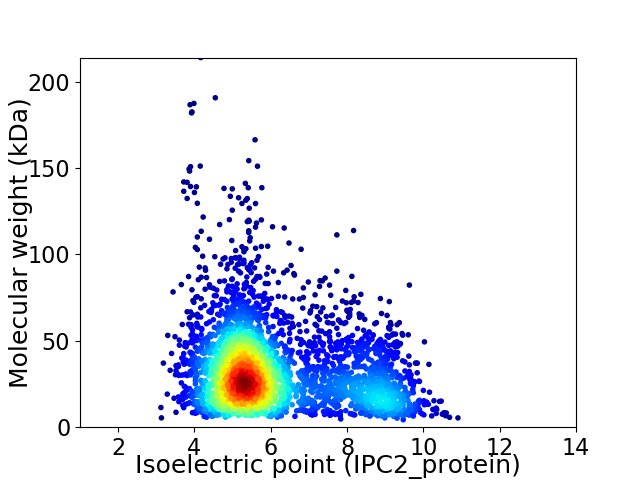
Virtual 2D-PAGE plot for 3501 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4WB73|A0A1Y4WB73_9FIRM Uncharacterized protein OS=Flavonifractor sp. An10 OX=1965537 GN=B5E42_16925 PE=4 SV=1
MM1 pKa = 7.05EE2 pKa = 4.52TFVSNFRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.57SKK14 pKa = 9.36MRR16 pKa = 11.84NLKK19 pKa = 8.77RR20 pKa = 11.84TLSLVLAVVMVIGLMVVGASAVSYY44 pKa = 11.34NDD46 pKa = 3.04FSDD49 pKa = 4.22RR50 pKa = 11.84EE51 pKa = 4.17EE52 pKa = 4.31IVNKK56 pKa = 10.34DD57 pKa = 3.57AVSMLTTLGIIEE69 pKa = 4.31GQPDD73 pKa = 3.75GSYY76 pKa = 11.27NPTGDD81 pKa = 3.1VDD83 pKa = 3.94RR84 pKa = 11.84AQMAKK89 pKa = 9.16MISVALTNNEE99 pKa = 4.29DD100 pKa = 4.21CDD102 pKa = 4.02TLYY105 pKa = 11.16TNVNSGLTDD114 pKa = 3.46ITANWARR121 pKa = 11.84GYY123 pKa = 10.58INYY126 pKa = 9.77CYY128 pKa = 10.64VRR130 pKa = 11.84GIIAGRR136 pKa = 11.84GDD138 pKa = 3.45NTFDD142 pKa = 3.89PDD144 pKa = 3.84ANVTGVEE151 pKa = 3.96AAKK154 pKa = 9.98MLLAALGYY162 pKa = 10.05NADD165 pKa = 3.52IEE167 pKa = 4.77GLVGPDD173 pKa = 2.83WALNTAALAQQLGIFRR189 pKa = 11.84NFTKK193 pKa = 10.53DD194 pKa = 2.98VSEE197 pKa = 4.12PLSRR201 pKa = 11.84DD202 pKa = 3.27DD203 pKa = 5.17AALLIYY209 pKa = 10.43NALDD213 pKa = 3.21VEE215 pKa = 5.09MIQQYY220 pKa = 6.04QTGYY224 pKa = 8.82ALVYY228 pKa = 10.15ADD230 pKa = 4.58HH231 pKa = 6.62RR232 pKa = 11.84TILSSVFGVVRR243 pKa = 11.84VEE245 pKa = 4.35GVVTANEE252 pKa = 3.92WARR255 pKa = 11.84LEE257 pKa = 4.26EE258 pKa = 4.25TDD260 pKa = 3.79SDD262 pKa = 3.86AALRR266 pKa = 11.84TGKK269 pKa = 7.28TTLDD273 pKa = 3.37EE274 pKa = 4.35VVVYY278 pKa = 10.76DD279 pKa = 3.84STTSNTTVPEE289 pKa = 5.11GIRR292 pKa = 11.84EE293 pKa = 4.16DD294 pKa = 4.12DD295 pKa = 3.45PVTFNVSTPVDD306 pKa = 3.93MIGKK310 pKa = 9.0AVTMYY315 pKa = 10.71VEE317 pKa = 4.27KK318 pKa = 8.58TTILANSNVIGIATNDD334 pKa = 3.52EE335 pKa = 4.41ANVIHH340 pKa = 6.46ATAATEE346 pKa = 4.03EE347 pKa = 4.31TLDD350 pKa = 4.72DD351 pKa = 4.72LLDD354 pKa = 3.88GTGIEE359 pKa = 4.47TDD361 pKa = 3.65RR362 pKa = 11.84NTQYY366 pKa = 10.89YY367 pKa = 9.68VNYY370 pKa = 9.16GWCEE374 pKa = 3.94DD375 pKa = 3.66GATEE379 pKa = 4.81AVEE382 pKa = 6.05LINQYY387 pKa = 10.75PDD389 pKa = 3.08AQLEE393 pKa = 4.37DD394 pKa = 4.56KK395 pKa = 11.32DD396 pKa = 3.63PGYY399 pKa = 11.27YY400 pKa = 10.14SLNGVEE406 pKa = 5.64VEE408 pKa = 4.9VIDD411 pKa = 5.16NNDD414 pKa = 3.74DD415 pKa = 3.42GTAEE419 pKa = 4.08YY420 pKa = 11.09VLYY423 pKa = 10.5LQEE426 pKa = 4.25TLSEE430 pKa = 4.15VTRR433 pKa = 11.84YY434 pKa = 9.8SDD436 pKa = 3.52RR437 pKa = 11.84NEE439 pKa = 4.19TISFYY444 pKa = 10.59IPEE447 pKa = 4.98LNNSGAPTGDD457 pKa = 3.26SDD459 pKa = 4.03TVTRR463 pKa = 11.84DD464 pKa = 3.28FEE466 pKa = 5.03DD467 pKa = 4.11VVFNDD472 pKa = 5.05DD473 pKa = 3.35VAADD477 pKa = 3.92DD478 pKa = 4.97LILYY482 pKa = 9.36VEE484 pKa = 4.38YY485 pKa = 10.75GGRR488 pKa = 11.84TYY490 pKa = 10.64IQLAPIVTDD499 pKa = 2.87TMNRR503 pKa = 11.84VDD505 pKa = 4.23RR506 pKa = 11.84DD507 pKa = 3.63RR508 pKa = 11.84DD509 pKa = 3.57SEE511 pKa = 4.9LYY513 pKa = 8.65ITLDD517 pKa = 2.83NGEE520 pKa = 4.91EE521 pKa = 3.92YY522 pKa = 10.41RR523 pKa = 11.84QAYY526 pKa = 9.64LPDD529 pKa = 4.31AASMTDD535 pKa = 3.16VEE537 pKa = 4.83LTHH540 pKa = 7.25FDD542 pKa = 3.32IDD544 pKa = 3.6NARR547 pKa = 11.84ADD549 pKa = 3.26IGFDD553 pKa = 3.42DD554 pKa = 5.68AYY556 pKa = 11.21DD557 pKa = 4.89FILDD561 pKa = 3.44ATEE564 pKa = 4.02QYY566 pKa = 10.82VIAVRR571 pKa = 11.84PAEE574 pKa = 4.11EE575 pKa = 3.85QVTNYY580 pKa = 11.27ALVLGSAWTQNALTRR595 pKa = 11.84SGEE598 pKa = 4.22VKK600 pKa = 10.17ILMADD605 pKa = 3.5GTEE608 pKa = 3.91NTFDD612 pKa = 3.85IDD614 pKa = 3.35WNEE617 pKa = 3.74SRR619 pKa = 11.84KK620 pKa = 9.83VFEE623 pKa = 4.79NEE625 pKa = 3.7TTNTRR630 pKa = 11.84KK631 pKa = 8.78DD632 pKa = 3.43TALEE636 pKa = 4.25NYY638 pKa = 10.17LGTRR642 pKa = 11.84DD643 pKa = 3.77VQHH646 pKa = 6.48SGSYY650 pKa = 8.36RR651 pKa = 11.84TGAAVGTVIEE661 pKa = 4.54YY662 pKa = 10.77SLNDD666 pKa = 3.84AGDD669 pKa = 3.54EE670 pKa = 4.01LTIEE674 pKa = 4.86NILNLNTLADD684 pKa = 3.82GGYY687 pKa = 10.27VNQNNGDD694 pKa = 3.99PLVDD698 pKa = 3.67VVSNPIAYY706 pKa = 9.19IAANRR711 pKa = 11.84AASDD715 pKa = 3.56TSVRR719 pKa = 11.84TANNLQYY726 pKa = 10.29TIQTGQPYY734 pKa = 8.59STGDD738 pKa = 2.99GTLYY742 pKa = 9.96LTVKK746 pKa = 9.32NTDD749 pKa = 2.98AVADD753 pKa = 3.87ARR755 pKa = 11.84NDD757 pKa = 3.58NASFAIDD764 pKa = 3.5RR765 pKa = 11.84DD766 pKa = 4.19TIAFYY771 pKa = 11.11YY772 pKa = 10.28VDD774 pKa = 3.56SDD776 pKa = 4.12TYY778 pKa = 11.47GVATGYY784 pKa = 11.39SNMSDD789 pKa = 3.41IPVQDD794 pKa = 3.45EE795 pKa = 4.19DD796 pKa = 4.34GNPVSVQVYY805 pKa = 8.97PVLEE809 pKa = 4.31KK810 pKa = 11.01NNQGGWTASRR820 pKa = 11.84LADD823 pKa = 3.21VALFNSEE830 pKa = 4.44PSNDD834 pKa = 3.63SNDD837 pKa = 3.51YY838 pKa = 10.41MLVLNANYY846 pKa = 10.2YY847 pKa = 10.59RR848 pKa = 11.84GDD850 pKa = 3.97EE851 pKa = 3.89LWLNVVFEE859 pKa = 5.65DD860 pKa = 3.99GTAATIEE867 pKa = 3.92IDD869 pKa = 4.63DD870 pKa = 4.85DD871 pKa = 4.27GGHH874 pKa = 5.97NFDD877 pKa = 4.09RR878 pKa = 11.84TSSYY882 pKa = 9.36MKK884 pKa = 10.15AYY886 pKa = 10.25AYY888 pKa = 10.63VEE890 pKa = 4.4NADD893 pKa = 3.46GTYY896 pKa = 10.19DD897 pKa = 3.48ISDD900 pKa = 3.7RR901 pKa = 11.84DD902 pKa = 3.92PIGEE906 pKa = 4.06YY907 pKa = 10.08DD908 pKa = 3.97AHH910 pKa = 6.04RR911 pKa = 11.84TDD913 pKa = 3.7NGTIYY918 pKa = 9.88WAAPTSVSPNPYY930 pKa = 8.74LTLLDD935 pKa = 4.41SSNVWDD941 pKa = 3.89VTDD944 pKa = 3.65VDD946 pKa = 4.6SANDD950 pKa = 4.67DD951 pKa = 3.28ITAGTFTRR959 pKa = 11.84TDD961 pKa = 3.16KK962 pKa = 11.19NAVIVTGGSRR972 pKa = 11.84DD973 pKa = 3.55EE974 pKa = 4.5TIRR977 pKa = 11.84AAWIWDD983 pKa = 3.39IDD985 pKa = 3.92DD986 pKa = 3.83TTVDD990 pKa = 4.88FDD992 pKa = 3.72TSDD995 pKa = 3.76YY996 pKa = 11.42LVVLSDD1002 pKa = 4.87ADD1004 pKa = 3.48NCAYY1008 pKa = 9.69IYY1010 pKa = 11.27YY1011 pKa = 10.35NDD1013 pKa = 5.06DD1014 pKa = 4.5LYY1016 pKa = 11.41DD1017 pKa = 4.16SPDD1020 pKa = 3.67FPDD1023 pKa = 5.5LDD1025 pKa = 3.9LCMSLIEE1032 pKa = 5.98AEE1034 pKa = 4.69MEE1036 pKa = 4.07DD1037 pKa = 3.24QGYY1040 pKa = 8.55TIVDD1044 pKa = 3.44RR1045 pKa = 11.84TRR1047 pKa = 11.84NGNTYY1052 pKa = 7.26TWEE1055 pKa = 4.19VEE1057 pKa = 4.11DD1058 pKa = 5.26SRR1060 pKa = 11.84GFDD1063 pKa = 3.0DD1064 pKa = 4.52TYY1066 pKa = 11.36RR1067 pKa = 11.84FVYY1070 pKa = 10.31DD1071 pKa = 3.33ATVSFTGNWLIPSYY1085 pKa = 10.38VGRR1088 pKa = 11.84VNGVEE1093 pKa = 4.23TFVPTASATITNRR1106 pKa = 11.84TDD1108 pKa = 2.9SSTHH1112 pKa = 5.87LAGSTPLAGTHH1123 pKa = 3.83VWGRR1127 pKa = 11.84LDD1129 pKa = 4.55ANNNYY1134 pKa = 10.4DD1135 pKa = 3.99WIAVGSVTVTGSGFDD1150 pKa = 3.37IEE1152 pKa = 5.17DD1153 pKa = 4.31GYY1155 pKa = 9.56TAVDD1159 pKa = 3.62VAGLALSITTDD1170 pKa = 2.89GDD1172 pKa = 3.67LDD1174 pKa = 3.8VAVTVNGDD1182 pKa = 3.71DD1183 pKa = 6.26LEE1185 pKa = 6.51DD1186 pKa = 4.98CDD1188 pKa = 5.22TYY1190 pKa = 11.66DD1191 pKa = 3.22AANDD1195 pKa = 3.54VVYY1198 pKa = 11.21VEE1200 pKa = 5.57GNDD1203 pKa = 3.51NEE1205 pKa = 4.38IVATVTGSFEE1215 pKa = 3.7ITTASDD1221 pKa = 3.06TKK1223 pKa = 11.07YY1224 pKa = 10.46IAGTNTNGLVRR1235 pKa = 11.84DD1236 pKa = 3.78VAGYY1240 pKa = 9.38DD1241 pKa = 3.31ASRR1244 pKa = 11.84NNATVTTPTGSGQIVFTASADD1265 pKa = 3.53GATTFEE1271 pKa = 4.34DD1272 pKa = 4.08VEE1274 pKa = 4.87VTFTWTVDD1282 pKa = 3.52QQTTTVDD1289 pKa = 3.46LNLATDD1295 pKa = 4.26LTT1297 pKa = 4.35
MM1 pKa = 7.05EE2 pKa = 4.52TFVSNFRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.57SKK14 pKa = 9.36MRR16 pKa = 11.84NLKK19 pKa = 8.77RR20 pKa = 11.84TLSLVLAVVMVIGLMVVGASAVSYY44 pKa = 11.34NDD46 pKa = 3.04FSDD49 pKa = 4.22RR50 pKa = 11.84EE51 pKa = 4.17EE52 pKa = 4.31IVNKK56 pKa = 10.34DD57 pKa = 3.57AVSMLTTLGIIEE69 pKa = 4.31GQPDD73 pKa = 3.75GSYY76 pKa = 11.27NPTGDD81 pKa = 3.1VDD83 pKa = 3.94RR84 pKa = 11.84AQMAKK89 pKa = 9.16MISVALTNNEE99 pKa = 4.29DD100 pKa = 4.21CDD102 pKa = 4.02TLYY105 pKa = 11.16TNVNSGLTDD114 pKa = 3.46ITANWARR121 pKa = 11.84GYY123 pKa = 10.58INYY126 pKa = 9.77CYY128 pKa = 10.64VRR130 pKa = 11.84GIIAGRR136 pKa = 11.84GDD138 pKa = 3.45NTFDD142 pKa = 3.89PDD144 pKa = 3.84ANVTGVEE151 pKa = 3.96AAKK154 pKa = 9.98MLLAALGYY162 pKa = 10.05NADD165 pKa = 3.52IEE167 pKa = 4.77GLVGPDD173 pKa = 2.83WALNTAALAQQLGIFRR189 pKa = 11.84NFTKK193 pKa = 10.53DD194 pKa = 2.98VSEE197 pKa = 4.12PLSRR201 pKa = 11.84DD202 pKa = 3.27DD203 pKa = 5.17AALLIYY209 pKa = 10.43NALDD213 pKa = 3.21VEE215 pKa = 5.09MIQQYY220 pKa = 6.04QTGYY224 pKa = 8.82ALVYY228 pKa = 10.15ADD230 pKa = 4.58HH231 pKa = 6.62RR232 pKa = 11.84TILSSVFGVVRR243 pKa = 11.84VEE245 pKa = 4.35GVVTANEE252 pKa = 3.92WARR255 pKa = 11.84LEE257 pKa = 4.26EE258 pKa = 4.25TDD260 pKa = 3.79SDD262 pKa = 3.86AALRR266 pKa = 11.84TGKK269 pKa = 7.28TTLDD273 pKa = 3.37EE274 pKa = 4.35VVVYY278 pKa = 10.76DD279 pKa = 3.84STTSNTTVPEE289 pKa = 5.11GIRR292 pKa = 11.84EE293 pKa = 4.16DD294 pKa = 4.12DD295 pKa = 3.45PVTFNVSTPVDD306 pKa = 3.93MIGKK310 pKa = 9.0AVTMYY315 pKa = 10.71VEE317 pKa = 4.27KK318 pKa = 8.58TTILANSNVIGIATNDD334 pKa = 3.52EE335 pKa = 4.41ANVIHH340 pKa = 6.46ATAATEE346 pKa = 4.03EE347 pKa = 4.31TLDD350 pKa = 4.72DD351 pKa = 4.72LLDD354 pKa = 3.88GTGIEE359 pKa = 4.47TDD361 pKa = 3.65RR362 pKa = 11.84NTQYY366 pKa = 10.89YY367 pKa = 9.68VNYY370 pKa = 9.16GWCEE374 pKa = 3.94DD375 pKa = 3.66GATEE379 pKa = 4.81AVEE382 pKa = 6.05LINQYY387 pKa = 10.75PDD389 pKa = 3.08AQLEE393 pKa = 4.37DD394 pKa = 4.56KK395 pKa = 11.32DD396 pKa = 3.63PGYY399 pKa = 11.27YY400 pKa = 10.14SLNGVEE406 pKa = 5.64VEE408 pKa = 4.9VIDD411 pKa = 5.16NNDD414 pKa = 3.74DD415 pKa = 3.42GTAEE419 pKa = 4.08YY420 pKa = 11.09VLYY423 pKa = 10.5LQEE426 pKa = 4.25TLSEE430 pKa = 4.15VTRR433 pKa = 11.84YY434 pKa = 9.8SDD436 pKa = 3.52RR437 pKa = 11.84NEE439 pKa = 4.19TISFYY444 pKa = 10.59IPEE447 pKa = 4.98LNNSGAPTGDD457 pKa = 3.26SDD459 pKa = 4.03TVTRR463 pKa = 11.84DD464 pKa = 3.28FEE466 pKa = 5.03DD467 pKa = 4.11VVFNDD472 pKa = 5.05DD473 pKa = 3.35VAADD477 pKa = 3.92DD478 pKa = 4.97LILYY482 pKa = 9.36VEE484 pKa = 4.38YY485 pKa = 10.75GGRR488 pKa = 11.84TYY490 pKa = 10.64IQLAPIVTDD499 pKa = 2.87TMNRR503 pKa = 11.84VDD505 pKa = 4.23RR506 pKa = 11.84DD507 pKa = 3.63RR508 pKa = 11.84DD509 pKa = 3.57SEE511 pKa = 4.9LYY513 pKa = 8.65ITLDD517 pKa = 2.83NGEE520 pKa = 4.91EE521 pKa = 3.92YY522 pKa = 10.41RR523 pKa = 11.84QAYY526 pKa = 9.64LPDD529 pKa = 4.31AASMTDD535 pKa = 3.16VEE537 pKa = 4.83LTHH540 pKa = 7.25FDD542 pKa = 3.32IDD544 pKa = 3.6NARR547 pKa = 11.84ADD549 pKa = 3.26IGFDD553 pKa = 3.42DD554 pKa = 5.68AYY556 pKa = 11.21DD557 pKa = 4.89FILDD561 pKa = 3.44ATEE564 pKa = 4.02QYY566 pKa = 10.82VIAVRR571 pKa = 11.84PAEE574 pKa = 4.11EE575 pKa = 3.85QVTNYY580 pKa = 11.27ALVLGSAWTQNALTRR595 pKa = 11.84SGEE598 pKa = 4.22VKK600 pKa = 10.17ILMADD605 pKa = 3.5GTEE608 pKa = 3.91NTFDD612 pKa = 3.85IDD614 pKa = 3.35WNEE617 pKa = 3.74SRR619 pKa = 11.84KK620 pKa = 9.83VFEE623 pKa = 4.79NEE625 pKa = 3.7TTNTRR630 pKa = 11.84KK631 pKa = 8.78DD632 pKa = 3.43TALEE636 pKa = 4.25NYY638 pKa = 10.17LGTRR642 pKa = 11.84DD643 pKa = 3.77VQHH646 pKa = 6.48SGSYY650 pKa = 8.36RR651 pKa = 11.84TGAAVGTVIEE661 pKa = 4.54YY662 pKa = 10.77SLNDD666 pKa = 3.84AGDD669 pKa = 3.54EE670 pKa = 4.01LTIEE674 pKa = 4.86NILNLNTLADD684 pKa = 3.82GGYY687 pKa = 10.27VNQNNGDD694 pKa = 3.99PLVDD698 pKa = 3.67VVSNPIAYY706 pKa = 9.19IAANRR711 pKa = 11.84AASDD715 pKa = 3.56TSVRR719 pKa = 11.84TANNLQYY726 pKa = 10.29TIQTGQPYY734 pKa = 8.59STGDD738 pKa = 2.99GTLYY742 pKa = 9.96LTVKK746 pKa = 9.32NTDD749 pKa = 2.98AVADD753 pKa = 3.87ARR755 pKa = 11.84NDD757 pKa = 3.58NASFAIDD764 pKa = 3.5RR765 pKa = 11.84DD766 pKa = 4.19TIAFYY771 pKa = 11.11YY772 pKa = 10.28VDD774 pKa = 3.56SDD776 pKa = 4.12TYY778 pKa = 11.47GVATGYY784 pKa = 11.39SNMSDD789 pKa = 3.41IPVQDD794 pKa = 3.45EE795 pKa = 4.19DD796 pKa = 4.34GNPVSVQVYY805 pKa = 8.97PVLEE809 pKa = 4.31KK810 pKa = 11.01NNQGGWTASRR820 pKa = 11.84LADD823 pKa = 3.21VALFNSEE830 pKa = 4.44PSNDD834 pKa = 3.63SNDD837 pKa = 3.51YY838 pKa = 10.41MLVLNANYY846 pKa = 10.2YY847 pKa = 10.59RR848 pKa = 11.84GDD850 pKa = 3.97EE851 pKa = 3.89LWLNVVFEE859 pKa = 5.65DD860 pKa = 3.99GTAATIEE867 pKa = 3.92IDD869 pKa = 4.63DD870 pKa = 4.85DD871 pKa = 4.27GGHH874 pKa = 5.97NFDD877 pKa = 4.09RR878 pKa = 11.84TSSYY882 pKa = 9.36MKK884 pKa = 10.15AYY886 pKa = 10.25AYY888 pKa = 10.63VEE890 pKa = 4.4NADD893 pKa = 3.46GTYY896 pKa = 10.19DD897 pKa = 3.48ISDD900 pKa = 3.7RR901 pKa = 11.84DD902 pKa = 3.92PIGEE906 pKa = 4.06YY907 pKa = 10.08DD908 pKa = 3.97AHH910 pKa = 6.04RR911 pKa = 11.84TDD913 pKa = 3.7NGTIYY918 pKa = 9.88WAAPTSVSPNPYY930 pKa = 8.74LTLLDD935 pKa = 4.41SSNVWDD941 pKa = 3.89VTDD944 pKa = 3.65VDD946 pKa = 4.6SANDD950 pKa = 4.67DD951 pKa = 3.28ITAGTFTRR959 pKa = 11.84TDD961 pKa = 3.16KK962 pKa = 11.19NAVIVTGGSRR972 pKa = 11.84DD973 pKa = 3.55EE974 pKa = 4.5TIRR977 pKa = 11.84AAWIWDD983 pKa = 3.39IDD985 pKa = 3.92DD986 pKa = 3.83TTVDD990 pKa = 4.88FDD992 pKa = 3.72TSDD995 pKa = 3.76YY996 pKa = 11.42LVVLSDD1002 pKa = 4.87ADD1004 pKa = 3.48NCAYY1008 pKa = 9.69IYY1010 pKa = 11.27YY1011 pKa = 10.35NDD1013 pKa = 5.06DD1014 pKa = 4.5LYY1016 pKa = 11.41DD1017 pKa = 4.16SPDD1020 pKa = 3.67FPDD1023 pKa = 5.5LDD1025 pKa = 3.9LCMSLIEE1032 pKa = 5.98AEE1034 pKa = 4.69MEE1036 pKa = 4.07DD1037 pKa = 3.24QGYY1040 pKa = 8.55TIVDD1044 pKa = 3.44RR1045 pKa = 11.84TRR1047 pKa = 11.84NGNTYY1052 pKa = 7.26TWEE1055 pKa = 4.19VEE1057 pKa = 4.11DD1058 pKa = 5.26SRR1060 pKa = 11.84GFDD1063 pKa = 3.0DD1064 pKa = 4.52TYY1066 pKa = 11.36RR1067 pKa = 11.84FVYY1070 pKa = 10.31DD1071 pKa = 3.33ATVSFTGNWLIPSYY1085 pKa = 10.38VGRR1088 pKa = 11.84VNGVEE1093 pKa = 4.23TFVPTASATITNRR1106 pKa = 11.84TDD1108 pKa = 2.9SSTHH1112 pKa = 5.87LAGSTPLAGTHH1123 pKa = 3.83VWGRR1127 pKa = 11.84LDD1129 pKa = 4.55ANNNYY1134 pKa = 10.4DD1135 pKa = 3.99WIAVGSVTVTGSGFDD1150 pKa = 3.37IEE1152 pKa = 5.17DD1153 pKa = 4.31GYY1155 pKa = 9.56TAVDD1159 pKa = 3.62VAGLALSITTDD1170 pKa = 2.89GDD1172 pKa = 3.67LDD1174 pKa = 3.8VAVTVNGDD1182 pKa = 3.71DD1183 pKa = 6.26LEE1185 pKa = 6.51DD1186 pKa = 4.98CDD1188 pKa = 5.22TYY1190 pKa = 11.66DD1191 pKa = 3.22AANDD1195 pKa = 3.54VVYY1198 pKa = 11.21VEE1200 pKa = 5.57GNDD1203 pKa = 3.51NEE1205 pKa = 4.38IVATVTGSFEE1215 pKa = 3.7ITTASDD1221 pKa = 3.06TKK1223 pKa = 11.07YY1224 pKa = 10.46IAGTNTNGLVRR1235 pKa = 11.84DD1236 pKa = 3.78VAGYY1240 pKa = 9.38DD1241 pKa = 3.31ASRR1244 pKa = 11.84NNATVTTPTGSGQIVFTASADD1265 pKa = 3.53GATTFEE1271 pKa = 4.34DD1272 pKa = 4.08VEE1274 pKa = 4.87VTFTWTVDD1282 pKa = 3.52QQTTTVDD1289 pKa = 3.46LNLATDD1295 pKa = 4.26LTT1297 pKa = 4.35
Molecular weight: 141.97 kDa
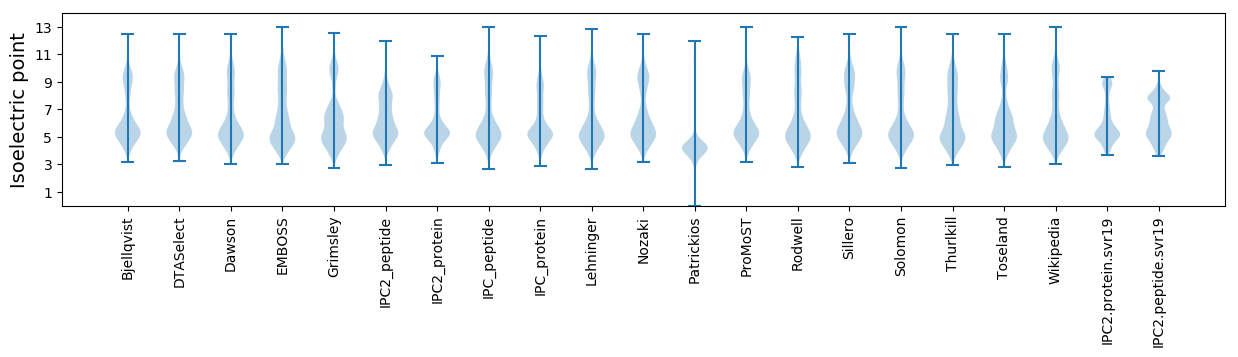
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4WG85|A0A1Y4WG85_9FIRM Uncharacterized protein OS=Flavonifractor sp. An10 OX=1965537 GN=B5E42_12325 PE=4 SV=1
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.24KK9 pKa = 7.68RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.13GFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MRR23 pKa = 11.84TANGRR28 pKa = 11.84KK29 pKa = 8.55VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.28
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.24KK9 pKa = 7.68RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.13GFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MRR23 pKa = 11.84TANGRR28 pKa = 11.84KK29 pKa = 8.55VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.28
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1082429 |
37 |
1867 |
309.2 |
34.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.147 ± 0.06 | 1.571 ± 0.017 |
5.609 ± 0.038 | 6.998 ± 0.043 |
3.683 ± 0.027 | 8.162 ± 0.04 |
1.851 ± 0.019 | 5.089 ± 0.044 |
4.165 ± 0.042 | 10.457 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.554 ± 0.02 | 3.042 ± 0.03 |
4.438 ± 0.031 | 3.479 ± 0.025 |
6.274 ± 0.048 | 5.253 ± 0.03 |
5.443 ± 0.038 | 7.134 ± 0.038 |
1.185 ± 0.017 | 3.468 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
