
Hepatitis B virus subtype adw2
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Orthohepadnavirus; Hepatitis B virus; Hepatitis B virus subtype adw
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
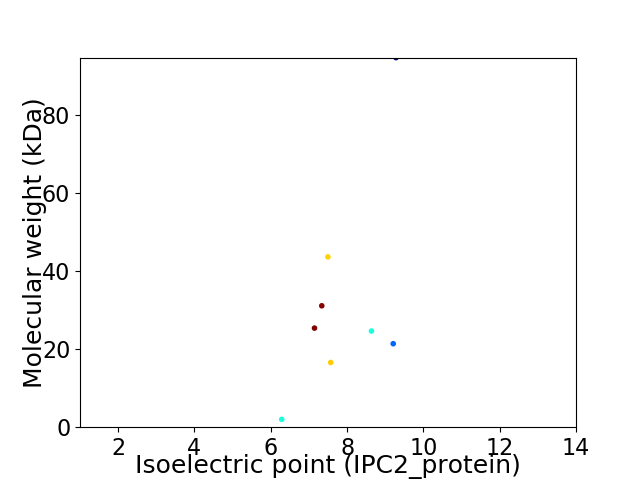
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
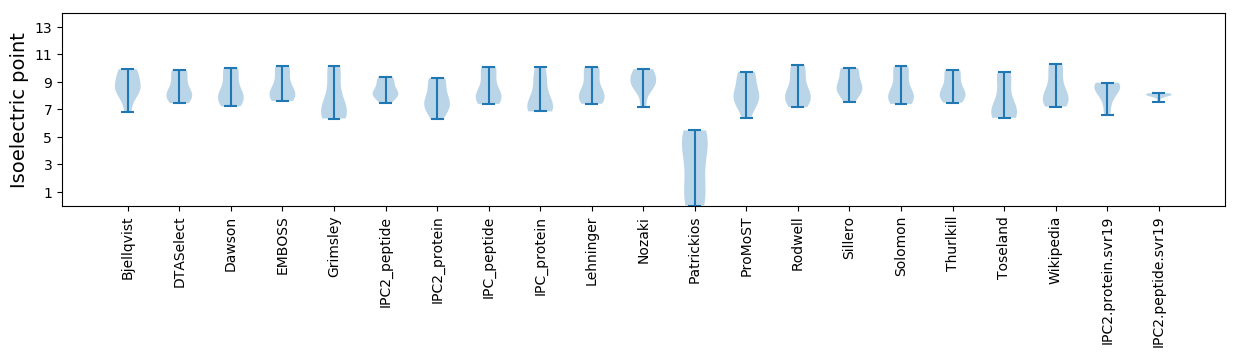
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q17UT4|Q17UT4_HBVAW Capsid protein OS=Hepatitis B virus subtype adw2 OX=10408 GN=pre C/C PE=3 SV=1
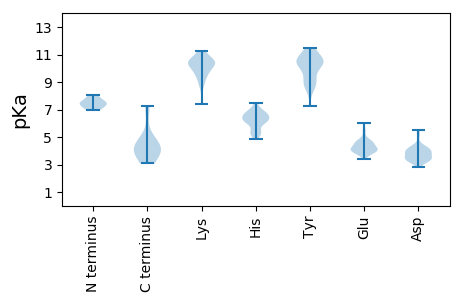
MM1 pKa = 7.1SHH3 pKa = 6.99CSSLQAVPWVALGHH17 pKa = 5.4GHH19 pKa = 7.28
MM1 pKa = 7.1SHH3 pKa = 6.99CSSLQAVPWVALGHH17 pKa = 5.4GHH19 pKa = 7.28
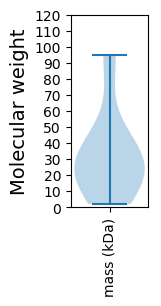
Molecular weight: 2.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q17UT5|Q17UT5_HBVAW Protein X OS=Hepatitis B virus subtype adw2 OX=10408 GN=X PE=3 SV=1
MM1 pKa = 7.31QLFHH5 pKa = 7.21LCLIISCTCPTVQASKK21 pKa = 11.2LCLGWLWGMDD31 pKa = 2.87IDD33 pKa = 4.46PYY35 pKa = 11.25KK36 pKa = 10.85EE37 pKa = 4.0FGATVEE43 pKa = 4.56LLSFLPSDD51 pKa = 4.16FFPSVRR57 pKa = 11.84DD58 pKa = 3.78LLDD61 pKa = 3.57TASALYY67 pKa = 10.4RR68 pKa = 11.84EE69 pKa = 4.55ALEE72 pKa = 4.62SPEE75 pKa = 4.18HH76 pKa = 6.65CSPHH80 pKa = 5.29HH81 pKa = 5.26TALRR85 pKa = 11.84QAILCWGEE93 pKa = 4.32LMTLATWVGNNLEE106 pKa = 4.75DD107 pKa = 3.84PASRR111 pKa = 11.84DD112 pKa = 3.34LVVNYY117 pKa = 10.33VNTNMGLKK125 pKa = 9.51IRR127 pKa = 11.84QLLWFHH133 pKa = 6.76ISCLTFGRR141 pKa = 11.84EE142 pKa = 3.77TVLEE146 pKa = 4.1YY147 pKa = 10.79LVSFGVWIRR156 pKa = 11.84TPPAYY161 pKa = 9.91RR162 pKa = 11.84PPNAPILSTLPEE174 pKa = 4.13TTVVRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84DD182 pKa = 3.21RR183 pKa = 11.84GRR185 pKa = 11.84SPRR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84TPSPRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84SQSPRR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84SQSRR210 pKa = 11.84EE211 pKa = 4.05SQCC214 pKa = 4.24
MM1 pKa = 7.31QLFHH5 pKa = 7.21LCLIISCTCPTVQASKK21 pKa = 11.2LCLGWLWGMDD31 pKa = 2.87IDD33 pKa = 4.46PYY35 pKa = 11.25KK36 pKa = 10.85EE37 pKa = 4.0FGATVEE43 pKa = 4.56LLSFLPSDD51 pKa = 4.16FFPSVRR57 pKa = 11.84DD58 pKa = 3.78LLDD61 pKa = 3.57TASALYY67 pKa = 10.4RR68 pKa = 11.84EE69 pKa = 4.55ALEE72 pKa = 4.62SPEE75 pKa = 4.18HH76 pKa = 6.65CSPHH80 pKa = 5.29HH81 pKa = 5.26TALRR85 pKa = 11.84QAILCWGEE93 pKa = 4.32LMTLATWVGNNLEE106 pKa = 4.75DD107 pKa = 3.84PASRR111 pKa = 11.84DD112 pKa = 3.34LVVNYY117 pKa = 10.33VNTNMGLKK125 pKa = 9.51IRR127 pKa = 11.84QLLWFHH133 pKa = 6.76ISCLTFGRR141 pKa = 11.84EE142 pKa = 3.77TVLEE146 pKa = 4.1YY147 pKa = 10.79LVSFGVWIRR156 pKa = 11.84TPPAYY161 pKa = 9.91RR162 pKa = 11.84PPNAPILSTLPEE174 pKa = 4.13TTVVRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84DD182 pKa = 3.21RR183 pKa = 11.84GRR185 pKa = 11.84SPRR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84TPSPRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84SQSPRR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84SQSRR210 pKa = 11.84EE211 pKa = 4.05SQCC214 pKa = 4.24
Molecular weight: 24.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2324 |
19 |
845 |
290.5 |
32.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.508 ± 0.581 | 3.657 ± 0.606 |
2.797 ± 0.31 | 2.453 ± 0.728 |
5.12 ± 0.43 | 6.54 ± 0.555 |
2.797 ± 0.673 | 4.561 ± 0.615 |
2.453 ± 0.565 | 11.833 ± 0.515 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.893 ± 0.258 | 3.485 ± 0.384 |
9.036 ± 0.756 | 3.744 ± 0.401 |
6.11 ± 1.419 | 10.327 ± 0.354 |
6.153 ± 0.538 | 5.637 ± 0.283 |
3.184 ± 0.63 | 2.711 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
