
Roseicitreum antarcticum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Roseicitreum
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
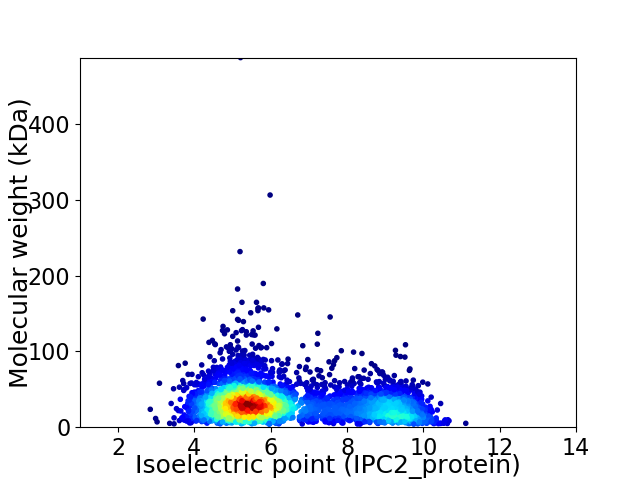
Virtual 2D-PAGE plot for 4002 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3BRP9|A0A1H3BRP9_9RHOB Lipoprotein-releasing system ATP-binding protein LolD OS=Roseicitreum antarcticum OX=564137 GN=lolD PE=3 SV=1
MM1 pKa = 6.29QTKK4 pKa = 10.09LYY6 pKa = 10.88AGVATLALIAGSASAQDD23 pKa = 3.67LVFGVGEE30 pKa = 4.17DD31 pKa = 4.78ARR33 pKa = 11.84MNWDD37 pKa = 3.44SYY39 pKa = 12.11NEE41 pKa = 3.75FAEE44 pKa = 4.82ANDD47 pKa = 3.9FSGQSVSLLGPWTGRR62 pKa = 11.84DD63 pKa = 3.39AEE65 pKa = 4.71LVNSTLAYY73 pKa = 9.76FAEE76 pKa = 4.51ATGATVNYY84 pKa = 9.53SGSDD88 pKa = 3.17SFEE91 pKa = 3.78QDD93 pKa = 2.48IVIAAQAGSAPNIAIFPQPGLAADD117 pKa = 4.14MASSGYY123 pKa = 10.63LSALDD128 pKa = 4.27EE129 pKa = 4.53STGDD133 pKa = 3.37WVRR136 pKa = 11.84EE137 pKa = 3.72NYY139 pKa = 10.1AAGEE143 pKa = 4.26SWVDD147 pKa = 3.17LGTYY151 pKa = 10.28AGADD155 pKa = 3.95GEE157 pKa = 4.49DD158 pKa = 3.43GLYY161 pKa = 11.04GFFYY165 pKa = 10.7KK166 pKa = 10.44VDD168 pKa = 3.56VKK170 pKa = 11.34SLVWYY175 pKa = 10.27NPEE178 pKa = 3.67QFFEE182 pKa = 4.22AGYY185 pKa = 10.01DD186 pKa = 3.32IPEE189 pKa = 4.18SMEE192 pKa = 4.05DD193 pKa = 3.43LKK195 pKa = 11.44ALSEE199 pKa = 4.19QIVADD204 pKa = 5.46GGTPWCLGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.2DD229 pKa = 3.44MMLRR233 pKa = 11.84LHH235 pKa = 6.33TPDD238 pKa = 5.38VYY240 pKa = 11.36DD241 pKa = 3.22GWVSNEE247 pKa = 3.62IAFDD251 pKa = 3.73DD252 pKa = 4.12PRR254 pKa = 11.84VVEE257 pKa = 4.96AIEE260 pKa = 4.44AYY262 pKa = 10.66GEE264 pKa = 3.96FSRR267 pKa = 11.84NNDD270 pKa = 2.79WVQGGSEE277 pKa = 3.97AAATTDD283 pKa = 4.27FRR285 pKa = 11.84DD286 pKa = 3.61SPGGLFSIPPACYY299 pKa = 8.41MHH301 pKa = 6.63KK302 pKa = 9.78QASFIPTFFPEE313 pKa = 4.11DD314 pKa = 3.27AEE316 pKa = 4.42YY317 pKa = 11.33GLDD320 pKa = 3.35YY321 pKa = 11.43DD322 pKa = 4.87FFYY325 pKa = 11.14FPASAEE331 pKa = 4.13ADD333 pKa = 3.16LGAPVLGAGTLFAITDD349 pKa = 3.75DD350 pKa = 3.88SDD352 pKa = 3.35ATRR355 pKa = 11.84GLMEE359 pKa = 4.42FLKK362 pKa = 10.27TPIAHH367 pKa = 7.05EE368 pKa = 3.41IWMAQSGFLTPHH380 pKa = 6.26TGVNSEE386 pKa = 4.42AYY388 pKa = 10.43GNDD391 pKa = 3.3SLRR394 pKa = 11.84AMGDD398 pKa = 2.82ILLNATTFRR407 pKa = 11.84FDD409 pKa = 5.1ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTGMVDD429 pKa = 3.45YY430 pKa = 7.64TTGADD435 pKa = 3.52ADD437 pKa = 4.31SVATTIQEE445 pKa = 3.6RR446 pKa = 11.84WNAMRR451 pKa = 5.25
MM1 pKa = 6.29QTKK4 pKa = 10.09LYY6 pKa = 10.88AGVATLALIAGSASAQDD23 pKa = 3.67LVFGVGEE30 pKa = 4.17DD31 pKa = 4.78ARR33 pKa = 11.84MNWDD37 pKa = 3.44SYY39 pKa = 12.11NEE41 pKa = 3.75FAEE44 pKa = 4.82ANDD47 pKa = 3.9FSGQSVSLLGPWTGRR62 pKa = 11.84DD63 pKa = 3.39AEE65 pKa = 4.71LVNSTLAYY73 pKa = 9.76FAEE76 pKa = 4.51ATGATVNYY84 pKa = 9.53SGSDD88 pKa = 3.17SFEE91 pKa = 3.78QDD93 pKa = 2.48IVIAAQAGSAPNIAIFPQPGLAADD117 pKa = 4.14MASSGYY123 pKa = 10.63LSALDD128 pKa = 4.27EE129 pKa = 4.53STGDD133 pKa = 3.37WVRR136 pKa = 11.84EE137 pKa = 3.72NYY139 pKa = 10.1AAGEE143 pKa = 4.26SWVDD147 pKa = 3.17LGTYY151 pKa = 10.28AGADD155 pKa = 3.95GEE157 pKa = 4.49DD158 pKa = 3.43GLYY161 pKa = 11.04GFFYY165 pKa = 10.7KK166 pKa = 10.44VDD168 pKa = 3.56VKK170 pKa = 11.34SLVWYY175 pKa = 10.27NPEE178 pKa = 3.67QFFEE182 pKa = 4.22AGYY185 pKa = 10.01DD186 pKa = 3.32IPEE189 pKa = 4.18SMEE192 pKa = 4.05DD193 pKa = 3.43LKK195 pKa = 11.44ALSEE199 pKa = 4.19QIVADD204 pKa = 5.46GGTPWCLGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.2DD229 pKa = 3.44MMLRR233 pKa = 11.84LHH235 pKa = 6.33TPDD238 pKa = 5.38VYY240 pKa = 11.36DD241 pKa = 3.22GWVSNEE247 pKa = 3.62IAFDD251 pKa = 3.73DD252 pKa = 4.12PRR254 pKa = 11.84VVEE257 pKa = 4.96AIEE260 pKa = 4.44AYY262 pKa = 10.66GEE264 pKa = 3.96FSRR267 pKa = 11.84NNDD270 pKa = 2.79WVQGGSEE277 pKa = 3.97AAATTDD283 pKa = 4.27FRR285 pKa = 11.84DD286 pKa = 3.61SPGGLFSIPPACYY299 pKa = 8.41MHH301 pKa = 6.63KK302 pKa = 9.78QASFIPTFFPEE313 pKa = 4.11DD314 pKa = 3.27AEE316 pKa = 4.42YY317 pKa = 11.33GLDD320 pKa = 3.35YY321 pKa = 11.43DD322 pKa = 4.87FFYY325 pKa = 11.14FPASAEE331 pKa = 4.13ADD333 pKa = 3.16LGAPVLGAGTLFAITDD349 pKa = 3.75DD350 pKa = 3.88SDD352 pKa = 3.35ATRR355 pKa = 11.84GLMEE359 pKa = 4.42FLKK362 pKa = 10.27TPIAHH367 pKa = 7.05EE368 pKa = 3.41IWMAQSGFLTPHH380 pKa = 6.26TGVNSEE386 pKa = 4.42AYY388 pKa = 10.43GNDD391 pKa = 3.3SLRR394 pKa = 11.84AMGDD398 pKa = 2.82ILLNATTFRR407 pKa = 11.84FDD409 pKa = 5.1ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTGMVDD429 pKa = 3.45YY430 pKa = 7.64TTGADD435 pKa = 3.52ADD437 pKa = 4.31SVATTIQEE445 pKa = 3.6RR446 pKa = 11.84WNAMRR451 pKa = 5.25
Molecular weight: 48.5 kDa
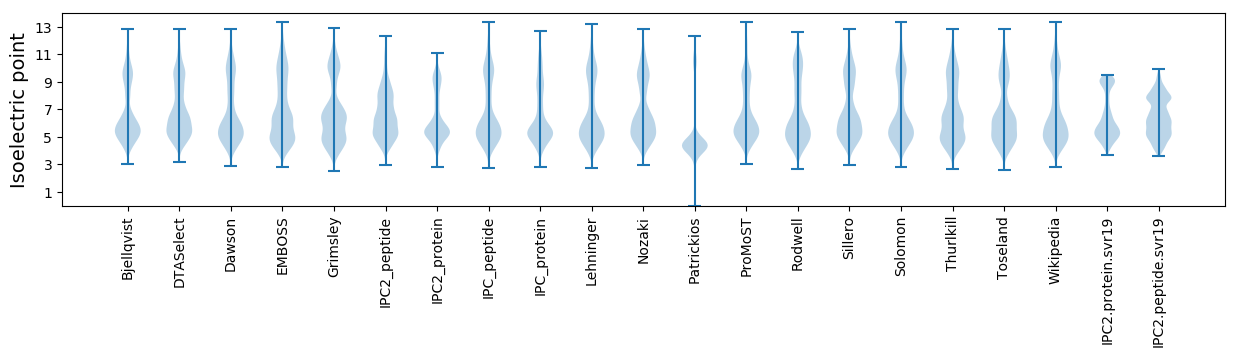
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2X6D9|A0A1H2X6D9_9RHOB Ribonuclease P protein component OS=Roseicitreum antarcticum OX=564137 GN=rnpA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27AGQKK29 pKa = 9.71IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27AGQKK29 pKa = 9.71IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1234146 |
39 |
4479 |
308.4 |
33.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.333 ± 0.055 | 0.864 ± 0.012 |
6.092 ± 0.031 | 5.02 ± 0.041 |
3.564 ± 0.026 | 8.774 ± 0.036 |
2.123 ± 0.02 | 4.995 ± 0.034 |
2.512 ± 0.03 | 10.13 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.857 ± 0.021 | 2.414 ± 0.019 |
5.371 ± 0.031 | 3.382 ± 0.021 |
7.106 ± 0.039 | 4.978 ± 0.026 |
5.692 ± 0.027 | 7.326 ± 0.034 |
1.397 ± 0.017 | 2.069 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
