
Rhinolophus ferrumequinum circovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; Bat associated circovirus 3
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins
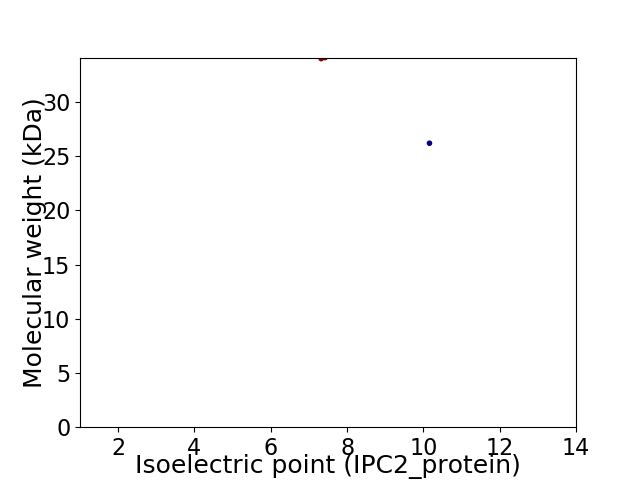
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3VR57|I3VR57_9CIRC Cap OS=Rhinolophus ferrumequinum circovirus 1 OX=1195373 PE=3 SV=1
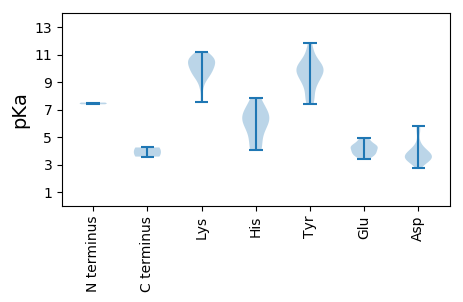
MM1 pKa = 7.51KK2 pKa = 10.36GDD4 pKa = 3.67EE5 pKa = 4.41TPGRR9 pKa = 11.84RR10 pKa = 11.84WCFTINNYY18 pKa = 7.72DD19 pKa = 3.88TPDD22 pKa = 3.2LTAVNEE28 pKa = 4.06AFKK31 pKa = 11.03EE32 pKa = 3.95KK33 pKa = 10.69DD34 pKa = 3.03VVYY37 pKa = 9.99AVCGKK42 pKa = 10.01EE43 pKa = 3.91VGKK46 pKa = 10.77KK47 pKa = 7.54GTKK50 pKa = 9.12HH51 pKa = 5.1LQGFIHH57 pKa = 6.42FTGNWRR63 pKa = 11.84FNRR66 pKa = 11.84VKK68 pKa = 10.72SLLGGRR74 pKa = 11.84AHH76 pKa = 7.04IEE78 pKa = 3.74KK79 pKa = 10.69ARR81 pKa = 11.84GNDD84 pKa = 3.48EE85 pKa = 4.02QNKK88 pKa = 8.92VYY90 pKa = 9.14CTKK93 pKa = 10.73EE94 pKa = 3.58EE95 pKa = 4.34TYY97 pKa = 11.11LEE99 pKa = 4.28VGSPQFQGKK108 pKa = 10.15RR109 pKa = 11.84NDD111 pKa = 3.29LGRR114 pKa = 11.84AVAALEE120 pKa = 4.72GGSSLSEE127 pKa = 3.8VAQANPEE134 pKa = 3.84VFIRR138 pKa = 11.84YY139 pKa = 8.7GRR141 pKa = 11.84GLRR144 pKa = 11.84DD145 pKa = 3.38YY146 pKa = 9.8MNVRR150 pKa = 11.84GLVKK154 pKa = 10.22PRR156 pKa = 11.84DD157 pKa = 3.98FKK159 pKa = 9.91THH161 pKa = 6.04VIVLVGEE168 pKa = 4.32PGSGKK173 pKa = 10.11SKK175 pKa = 10.16YY176 pKa = 9.83ANEE179 pKa = 4.27VEE181 pKa = 4.35GTKK184 pKa = 9.81YY185 pKa = 9.4WKK187 pKa = 9.87PRR189 pKa = 11.84GQWWDD194 pKa = 3.5GYY196 pKa = 10.31NGEE199 pKa = 4.45EE200 pKa = 4.84VVVLDD205 pKa = 5.81DD206 pKa = 4.04FYY208 pKa = 11.81GWVPYY213 pKa = 10.66DD214 pKa = 3.32EE215 pKa = 4.89LLRR218 pKa = 11.84VGDD221 pKa = 4.41RR222 pKa = 11.84YY223 pKa = 9.98PLKK226 pKa = 10.61VQVKK230 pKa = 8.45GAFVDD235 pKa = 5.31FVSKK239 pKa = 9.61TLVITSNKK247 pKa = 9.45RR248 pKa = 11.84PEE250 pKa = 3.55EE251 pKa = 4.22WYY253 pKa = 10.19DD254 pKa = 3.32KK255 pKa = 11.21EE256 pKa = 4.84KK257 pKa = 10.93IPDD260 pKa = 3.49QSAMWRR266 pKa = 11.84RR267 pKa = 11.84FNEE270 pKa = 3.61MYY272 pKa = 9.76YY273 pKa = 10.17CEE275 pKa = 4.48RR276 pKa = 11.84GEE278 pKa = 4.23PIKK281 pKa = 10.21PYY283 pKa = 9.16PDD285 pKa = 2.78EE286 pKa = 4.1WKK288 pKa = 10.66EE289 pKa = 3.76FEE291 pKa = 4.56TNFF294 pKa = 4.24
MM1 pKa = 7.51KK2 pKa = 10.36GDD4 pKa = 3.67EE5 pKa = 4.41TPGRR9 pKa = 11.84RR10 pKa = 11.84WCFTINNYY18 pKa = 7.72DD19 pKa = 3.88TPDD22 pKa = 3.2LTAVNEE28 pKa = 4.06AFKK31 pKa = 11.03EE32 pKa = 3.95KK33 pKa = 10.69DD34 pKa = 3.03VVYY37 pKa = 9.99AVCGKK42 pKa = 10.01EE43 pKa = 3.91VGKK46 pKa = 10.77KK47 pKa = 7.54GTKK50 pKa = 9.12HH51 pKa = 5.1LQGFIHH57 pKa = 6.42FTGNWRR63 pKa = 11.84FNRR66 pKa = 11.84VKK68 pKa = 10.72SLLGGRR74 pKa = 11.84AHH76 pKa = 7.04IEE78 pKa = 3.74KK79 pKa = 10.69ARR81 pKa = 11.84GNDD84 pKa = 3.48EE85 pKa = 4.02QNKK88 pKa = 8.92VYY90 pKa = 9.14CTKK93 pKa = 10.73EE94 pKa = 3.58EE95 pKa = 4.34TYY97 pKa = 11.11LEE99 pKa = 4.28VGSPQFQGKK108 pKa = 10.15RR109 pKa = 11.84NDD111 pKa = 3.29LGRR114 pKa = 11.84AVAALEE120 pKa = 4.72GGSSLSEE127 pKa = 3.8VAQANPEE134 pKa = 3.84VFIRR138 pKa = 11.84YY139 pKa = 8.7GRR141 pKa = 11.84GLRR144 pKa = 11.84DD145 pKa = 3.38YY146 pKa = 9.8MNVRR150 pKa = 11.84GLVKK154 pKa = 10.22PRR156 pKa = 11.84DD157 pKa = 3.98FKK159 pKa = 9.91THH161 pKa = 6.04VIVLVGEE168 pKa = 4.32PGSGKK173 pKa = 10.11SKK175 pKa = 10.16YY176 pKa = 9.83ANEE179 pKa = 4.27VEE181 pKa = 4.35GTKK184 pKa = 9.81YY185 pKa = 9.4WKK187 pKa = 9.87PRR189 pKa = 11.84GQWWDD194 pKa = 3.5GYY196 pKa = 10.31NGEE199 pKa = 4.45EE200 pKa = 4.84VVVLDD205 pKa = 5.81DD206 pKa = 4.04FYY208 pKa = 11.81GWVPYY213 pKa = 10.66DD214 pKa = 3.32EE215 pKa = 4.89LLRR218 pKa = 11.84VGDD221 pKa = 4.41RR222 pKa = 11.84YY223 pKa = 9.98PLKK226 pKa = 10.61VQVKK230 pKa = 8.45GAFVDD235 pKa = 5.31FVSKK239 pKa = 9.61TLVITSNKK247 pKa = 9.45RR248 pKa = 11.84PEE250 pKa = 3.55EE251 pKa = 4.22WYY253 pKa = 10.19DD254 pKa = 3.32KK255 pKa = 11.21EE256 pKa = 4.84KK257 pKa = 10.93IPDD260 pKa = 3.49QSAMWRR266 pKa = 11.84RR267 pKa = 11.84FNEE270 pKa = 3.61MYY272 pKa = 9.76YY273 pKa = 10.17CEE275 pKa = 4.48RR276 pKa = 11.84GEE278 pKa = 4.23PIKK281 pKa = 10.21PYY283 pKa = 9.16PDD285 pKa = 2.78EE286 pKa = 4.1WKK288 pKa = 10.66EE289 pKa = 3.76FEE291 pKa = 4.56TNFF294 pKa = 4.24
Molecular weight: 34.0 kDa
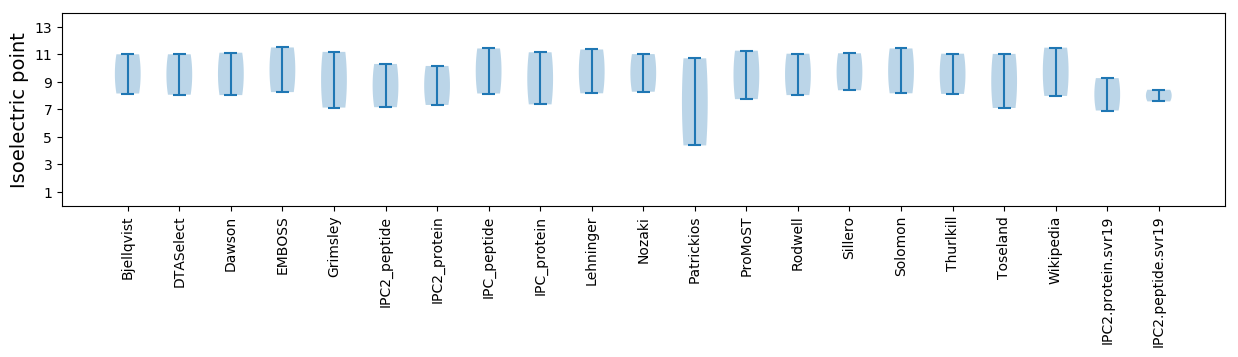
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3VR57|I3VR57_9CIRC Cap OS=Rhinolophus ferrumequinum circovirus 1 OX=1195373 PE=3 SV=1
MM1 pKa = 7.38PRR3 pKa = 11.84STRR6 pKa = 11.84HH7 pKa = 4.14RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84NQWFKK16 pKa = 10.53RR17 pKa = 11.84WRR19 pKa = 11.84QRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GHH26 pKa = 4.06TRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 7.92RR34 pKa = 11.84NKK36 pKa = 9.55VGIYY40 pKa = 9.0NFRR43 pKa = 11.84FRR45 pKa = 11.84AITTMTINKK54 pKa = 9.37NSNQGYY60 pKa = 7.57FTYY63 pKa = 9.91TLNGSVPTAFANYY76 pKa = 9.39FDD78 pKa = 4.52MYY80 pKa = 10.33RR81 pKa = 11.84IAKK84 pKa = 9.45VRR86 pKa = 11.84VQWLPMVSISEE97 pKa = 3.95VRR99 pKa = 11.84AWGATVIDD107 pKa = 3.75LTGRR111 pKa = 11.84DD112 pKa = 3.49TTVPSTGRR120 pKa = 11.84TDD122 pKa = 3.6FTIDD126 pKa = 3.33DD127 pKa = 3.77STRR130 pKa = 11.84RR131 pKa = 11.84LWNPTRR137 pKa = 11.84IHH139 pKa = 6.09SRR141 pKa = 11.84YY142 pKa = 7.42FTPKK146 pKa = 9.66PEE148 pKa = 3.6IQIRR152 pKa = 11.84SNSEE156 pKa = 3.39AVQPNNPRR164 pKa = 11.84NQLWIDD170 pKa = 3.55SRR172 pKa = 11.84DD173 pKa = 3.37KK174 pKa = 10.96DD175 pKa = 4.1VKK177 pKa = 10.29HH178 pKa = 6.6HH179 pKa = 6.39GIAYY183 pKa = 8.93YY184 pKa = 8.25FHH186 pKa = 7.83PDD188 pKa = 3.84DD189 pKa = 5.57IGDD192 pKa = 3.8DD193 pKa = 3.94VYY195 pKa = 11.34KK196 pKa = 10.57FSYY199 pKa = 9.79IVTYY203 pKa = 9.83YY204 pKa = 10.36FQFRR208 pKa = 11.84QFAGSQAPSVV218 pKa = 3.58
MM1 pKa = 7.38PRR3 pKa = 11.84STRR6 pKa = 11.84HH7 pKa = 4.14RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84NQWFKK16 pKa = 10.53RR17 pKa = 11.84WRR19 pKa = 11.84QRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GHH26 pKa = 4.06TRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 7.92RR34 pKa = 11.84NKK36 pKa = 9.55VGIYY40 pKa = 9.0NFRR43 pKa = 11.84FRR45 pKa = 11.84AITTMTINKK54 pKa = 9.37NSNQGYY60 pKa = 7.57FTYY63 pKa = 9.91TLNGSVPTAFANYY76 pKa = 9.39FDD78 pKa = 4.52MYY80 pKa = 10.33RR81 pKa = 11.84IAKK84 pKa = 9.45VRR86 pKa = 11.84VQWLPMVSISEE97 pKa = 3.95VRR99 pKa = 11.84AWGATVIDD107 pKa = 3.75LTGRR111 pKa = 11.84DD112 pKa = 3.49TTVPSTGRR120 pKa = 11.84TDD122 pKa = 3.6FTIDD126 pKa = 3.33DD127 pKa = 3.77STRR130 pKa = 11.84RR131 pKa = 11.84LWNPTRR137 pKa = 11.84IHH139 pKa = 6.09SRR141 pKa = 11.84YY142 pKa = 7.42FTPKK146 pKa = 9.66PEE148 pKa = 3.6IQIRR152 pKa = 11.84SNSEE156 pKa = 3.39AVQPNNPRR164 pKa = 11.84NQLWIDD170 pKa = 3.55SRR172 pKa = 11.84DD173 pKa = 3.37KK174 pKa = 10.96DD175 pKa = 4.1VKK177 pKa = 10.29HH178 pKa = 6.6HH179 pKa = 6.39GIAYY183 pKa = 8.93YY184 pKa = 8.25FHH186 pKa = 7.83PDD188 pKa = 3.84DD189 pKa = 5.57IGDD192 pKa = 3.8DD193 pKa = 3.94VYY195 pKa = 11.34KK196 pKa = 10.57FSYY199 pKa = 9.79IVTYY203 pKa = 9.83YY204 pKa = 10.36FQFRR208 pKa = 11.84QFAGSQAPSVV218 pKa = 3.58
Molecular weight: 26.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
512 |
218 |
294 |
256.0 |
30.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.492 ± 0.06 | 0.781 ± 0.497 |
5.859 ± 0.066 | 5.859 ± 2.853 |
5.273 ± 0.439 | 7.813 ± 1.76 |
1.953 ± 0.509 | 4.297 ± 1.352 |
6.641 ± 1.89 | 4.102 ± 1.15 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.563 ± 0.173 | 5.469 ± 0.315 |
4.883 ± 0.104 | 3.516 ± 0.682 |
9.961 ± 2.71 | 4.688 ± 1.104 |
6.25 ± 1.569 | 7.813 ± 1.177 |
3.125 ± 0.055 | 5.664 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
