
Changjiang tombus-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
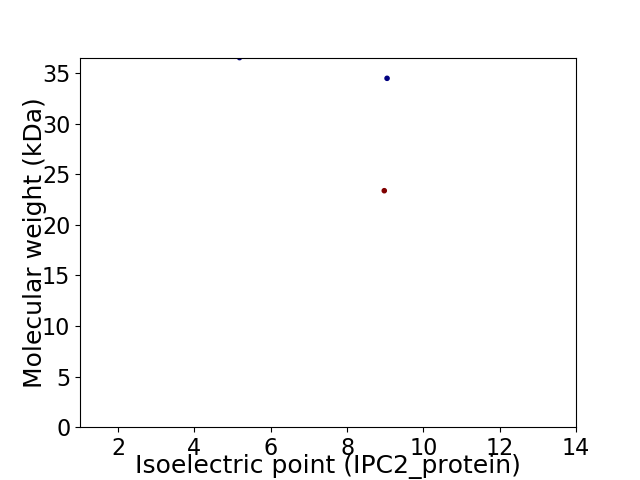
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KFY8|A0A1L3KFY8_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 11 OX=1922804 PE=4 SV=1
MM1 pKa = 7.62FKK3 pKa = 10.38RR4 pKa = 11.84AQTIRR9 pKa = 11.84QYY11 pKa = 10.29WSEE14 pKa = 3.85FKK16 pKa = 10.99KK17 pKa = 10.62PVFVGLDD24 pKa = 3.14ASRR27 pKa = 11.84FDD29 pKa = 3.3QHH31 pKa = 6.38VSRR34 pKa = 11.84EE35 pKa = 4.14ALEE38 pKa = 4.47FEE40 pKa = 4.5HH41 pKa = 7.13GLYY44 pKa = 10.08TEE46 pKa = 4.38VFRR49 pKa = 11.84DD50 pKa = 3.45PFLAYY55 pKa = 10.28LLNMQIDD62 pKa = 3.95QVGYY66 pKa = 11.17ANMADD71 pKa = 3.52GSVKK75 pKa = 10.76YY76 pKa = 10.4NVEE79 pKa = 3.86GCRR82 pKa = 11.84ASGDD86 pKa = 3.61MNTALGNVFLMCVITYY102 pKa = 10.09NYY104 pKa = 10.43LHH106 pKa = 7.23ALPCKK111 pKa = 9.37WRR113 pKa = 11.84FINDD117 pKa = 3.34GDD119 pKa = 3.88DD120 pKa = 3.34CGIFIEE126 pKa = 4.66HH127 pKa = 7.31ADD129 pKa = 3.43MHH131 pKa = 6.98LLDD134 pKa = 5.19EE135 pKa = 5.15LPSHH139 pKa = 6.42HH140 pKa = 6.88LLYY143 pKa = 10.58GFEE146 pKa = 4.57MEE148 pKa = 5.09VEE150 pKa = 4.09DD151 pKa = 3.85PVYY154 pKa = 10.26IIEE157 pKa = 4.37RR158 pKa = 11.84VEE160 pKa = 4.34FCQSRR165 pKa = 11.84PVQLNEE171 pKa = 4.21EE172 pKa = 4.04EE173 pKa = 4.0WMMVRR178 pKa = 11.84NIHH181 pKa = 6.72KK182 pKa = 10.15AIKK185 pKa = 9.84HH186 pKa = 5.56DD187 pKa = 4.54WICITSRR194 pKa = 11.84DD195 pKa = 3.49WATTEE200 pKa = 3.95EE201 pKa = 4.25NLVATSACGLALFGDD216 pKa = 4.46VPVLGPMYY224 pKa = 9.98QAMSRR229 pKa = 11.84FPHH232 pKa = 6.04RR233 pKa = 11.84QRR235 pKa = 11.84VVDD238 pKa = 4.75KK239 pKa = 11.07LLEE242 pKa = 4.36TKK244 pKa = 10.37EE245 pKa = 4.2GWRR248 pKa = 11.84SLLTGHH254 pKa = 7.4RR255 pKa = 11.84FNPVDD260 pKa = 3.48STIARR265 pKa = 11.84VSIYY269 pKa = 10.23KK270 pKa = 10.44AFGMLPDD277 pKa = 3.74EE278 pKa = 4.24QVEE281 pKa = 4.14LEE283 pKa = 4.16RR284 pKa = 11.84EE285 pKa = 4.05FRR287 pKa = 11.84AFDD290 pKa = 3.57PQNIKK295 pKa = 10.94DD296 pKa = 3.67KK297 pKa = 10.59TSLYY301 pKa = 8.77SQPSVRR307 pKa = 11.84VQYY310 pKa = 10.75IIDD313 pKa = 3.56TT314 pKa = 4.1
MM1 pKa = 7.62FKK3 pKa = 10.38RR4 pKa = 11.84AQTIRR9 pKa = 11.84QYY11 pKa = 10.29WSEE14 pKa = 3.85FKK16 pKa = 10.99KK17 pKa = 10.62PVFVGLDD24 pKa = 3.14ASRR27 pKa = 11.84FDD29 pKa = 3.3QHH31 pKa = 6.38VSRR34 pKa = 11.84EE35 pKa = 4.14ALEE38 pKa = 4.47FEE40 pKa = 4.5HH41 pKa = 7.13GLYY44 pKa = 10.08TEE46 pKa = 4.38VFRR49 pKa = 11.84DD50 pKa = 3.45PFLAYY55 pKa = 10.28LLNMQIDD62 pKa = 3.95QVGYY66 pKa = 11.17ANMADD71 pKa = 3.52GSVKK75 pKa = 10.76YY76 pKa = 10.4NVEE79 pKa = 3.86GCRR82 pKa = 11.84ASGDD86 pKa = 3.61MNTALGNVFLMCVITYY102 pKa = 10.09NYY104 pKa = 10.43LHH106 pKa = 7.23ALPCKK111 pKa = 9.37WRR113 pKa = 11.84FINDD117 pKa = 3.34GDD119 pKa = 3.88DD120 pKa = 3.34CGIFIEE126 pKa = 4.66HH127 pKa = 7.31ADD129 pKa = 3.43MHH131 pKa = 6.98LLDD134 pKa = 5.19EE135 pKa = 5.15LPSHH139 pKa = 6.42HH140 pKa = 6.88LLYY143 pKa = 10.58GFEE146 pKa = 4.57MEE148 pKa = 5.09VEE150 pKa = 4.09DD151 pKa = 3.85PVYY154 pKa = 10.26IIEE157 pKa = 4.37RR158 pKa = 11.84VEE160 pKa = 4.34FCQSRR165 pKa = 11.84PVQLNEE171 pKa = 4.21EE172 pKa = 4.04EE173 pKa = 4.0WMMVRR178 pKa = 11.84NIHH181 pKa = 6.72KK182 pKa = 10.15AIKK185 pKa = 9.84HH186 pKa = 5.56DD187 pKa = 4.54WICITSRR194 pKa = 11.84DD195 pKa = 3.49WATTEE200 pKa = 3.95EE201 pKa = 4.25NLVATSACGLALFGDD216 pKa = 4.46VPVLGPMYY224 pKa = 9.98QAMSRR229 pKa = 11.84FPHH232 pKa = 6.04RR233 pKa = 11.84QRR235 pKa = 11.84VVDD238 pKa = 4.75KK239 pKa = 11.07LLEE242 pKa = 4.36TKK244 pKa = 10.37EE245 pKa = 4.2GWRR248 pKa = 11.84SLLTGHH254 pKa = 7.4RR255 pKa = 11.84FNPVDD260 pKa = 3.48STIARR265 pKa = 11.84VSIYY269 pKa = 10.23KK270 pKa = 10.44AFGMLPDD277 pKa = 3.74EE278 pKa = 4.24QVEE281 pKa = 4.14LEE283 pKa = 4.16RR284 pKa = 11.84EE285 pKa = 4.05FRR287 pKa = 11.84AFDD290 pKa = 3.57PQNIKK295 pKa = 10.94DD296 pKa = 3.67KK297 pKa = 10.59TSLYY301 pKa = 8.77SQPSVRR307 pKa = 11.84VQYY310 pKa = 10.75IIDD313 pKa = 3.56TT314 pKa = 4.1
Molecular weight: 36.51 kDa
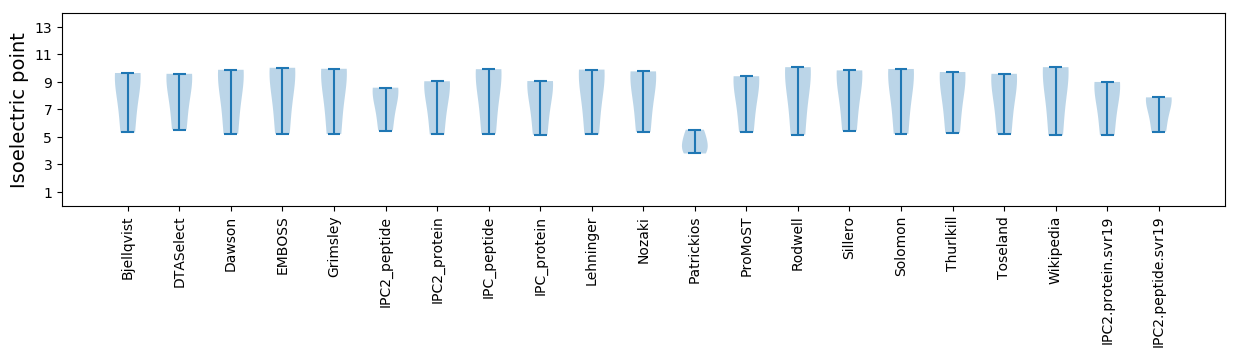
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFY8|A0A1L3KFY8_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 11 OX=1922804 PE=4 SV=1
MM1 pKa = 7.44ALEE4 pKa = 4.93IILLLVIAFEE14 pKa = 4.54IIVRR18 pKa = 11.84TKK20 pKa = 9.67VWDD23 pKa = 3.63AAGSLVTAALEE34 pKa = 4.52GCGRR38 pKa = 11.84AWTWMVPCFRR48 pKa = 11.84AILPILRR55 pKa = 11.84APILEE60 pKa = 4.33VVSATSCWVSVLYY73 pKa = 10.61HH74 pKa = 6.27SLLMGHH80 pKa = 6.47VRR82 pKa = 11.84RR83 pKa = 11.84QRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84IVLEE92 pKa = 3.76SSKK95 pKa = 9.69RR96 pKa = 11.84TQRR99 pKa = 11.84VLSVLDD105 pKa = 3.42MDD107 pKa = 5.67EE108 pKa = 5.21IGDD111 pKa = 4.35DD112 pKa = 3.63EE113 pKa = 4.54MEE115 pKa = 4.09LVGRR119 pKa = 11.84KK120 pKa = 9.63LYY122 pKa = 10.09VRR124 pKa = 11.84CAVLISRR131 pKa = 11.84RR132 pKa = 11.84ARR134 pKa = 11.84MGLKK138 pKa = 9.96YY139 pKa = 10.25PSYY142 pKa = 10.79SPANEE147 pKa = 4.15RR148 pKa = 11.84IAADD152 pKa = 4.37WILKK156 pKa = 9.25HH157 pKa = 6.62LPDD160 pKa = 5.01DD161 pKa = 3.5MTVGVRR167 pKa = 11.84HH168 pKa = 5.96KK169 pKa = 10.69VLPLAVKK176 pKa = 8.37LTFVRR181 pKa = 11.84SHH183 pKa = 7.25YY184 pKa = 9.61EE185 pKa = 3.71DD186 pKa = 3.56KK187 pKa = 11.15AALHH191 pKa = 5.33FQGLKK196 pKa = 10.16EE197 pKa = 3.9YY198 pKa = 11.3VDD200 pKa = 4.87FSRR203 pKa = 11.84HH204 pKa = 3.98
MM1 pKa = 7.44ALEE4 pKa = 4.93IILLLVIAFEE14 pKa = 4.54IIVRR18 pKa = 11.84TKK20 pKa = 9.67VWDD23 pKa = 3.63AAGSLVTAALEE34 pKa = 4.52GCGRR38 pKa = 11.84AWTWMVPCFRR48 pKa = 11.84AILPILRR55 pKa = 11.84APILEE60 pKa = 4.33VVSATSCWVSVLYY73 pKa = 10.61HH74 pKa = 6.27SLLMGHH80 pKa = 6.47VRR82 pKa = 11.84RR83 pKa = 11.84QRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84IVLEE92 pKa = 3.76SSKK95 pKa = 9.69RR96 pKa = 11.84TQRR99 pKa = 11.84VLSVLDD105 pKa = 3.42MDD107 pKa = 5.67EE108 pKa = 5.21IGDD111 pKa = 4.35DD112 pKa = 3.63EE113 pKa = 4.54MEE115 pKa = 4.09LVGRR119 pKa = 11.84KK120 pKa = 9.63LYY122 pKa = 10.09VRR124 pKa = 11.84CAVLISRR131 pKa = 11.84RR132 pKa = 11.84ARR134 pKa = 11.84MGLKK138 pKa = 9.96YY139 pKa = 10.25PSYY142 pKa = 10.79SPANEE147 pKa = 4.15RR148 pKa = 11.84IAADD152 pKa = 4.37WILKK156 pKa = 9.25HH157 pKa = 6.62LPDD160 pKa = 5.01DD161 pKa = 3.5MTVGVRR167 pKa = 11.84HH168 pKa = 5.96KK169 pKa = 10.69VLPLAVKK176 pKa = 8.37LTFVRR181 pKa = 11.84SHH183 pKa = 7.25YY184 pKa = 9.61EE185 pKa = 3.71DD186 pKa = 3.56KK187 pKa = 11.15AALHH191 pKa = 5.33FQGLKK196 pKa = 10.16EE197 pKa = 3.9YY198 pKa = 11.3VDD200 pKa = 4.87FSRR203 pKa = 11.84HH204 pKa = 3.98
Molecular weight: 23.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
841 |
204 |
323 |
280.3 |
31.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.037 ± 1.396 | 1.784 ± 0.229 |
4.875 ± 0.837 | 4.875 ± 1.335 |
4.162 ± 0.759 | 6.421 ± 1.018 |
2.973 ± 0.322 | 4.4 ± 0.898 |
3.805 ± 0.198 | 9.512 ± 0.992 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.092 ± 0.384 | 4.043 ± 1.178 |
4.637 ± 0.508 | 3.448 ± 0.616 |
6.659 ± 1.135 | 5.945 ± 0.372 |
6.302 ± 1.554 | 8.442 ± 0.728 |
1.902 ± 0.178 | 3.686 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
