
Howler monkey associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
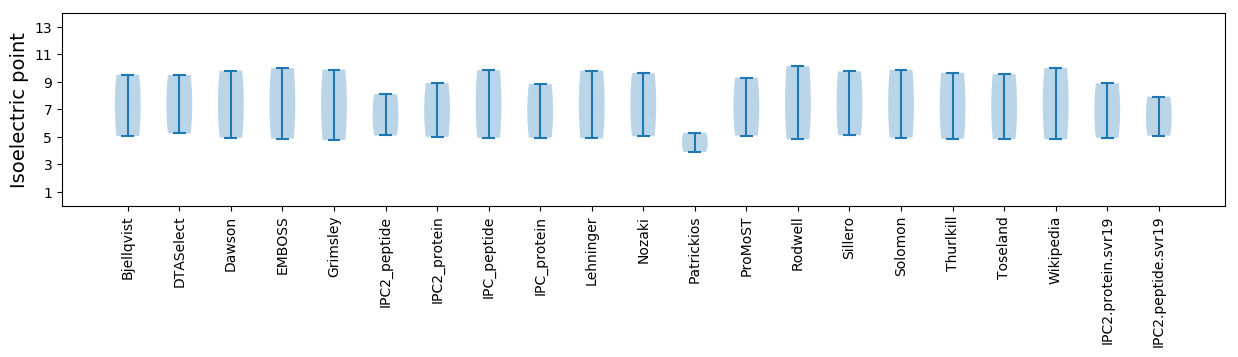
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0B5GP05|A0A0B5GP05_9VIRU Rep protein OS=Howler monkey associated porprismacovirus 1 OX=2170112 GN=rep PE=4 SV=1
MM1 pKa = 7.63GLRR4 pKa = 11.84RR5 pKa = 11.84NIPTAPMLTALWPALLGISLKK26 pKa = 10.61ARR28 pKa = 11.84NIFTDD33 pKa = 4.49FTSDD37 pKa = 3.17NMATNRR43 pKa = 11.84AYY45 pKa = 10.53ASFQEE50 pKa = 4.64IYY52 pKa = 10.64DD53 pKa = 3.36FHH55 pKa = 7.65TEE57 pKa = 3.83VGNTTLIEE65 pKa = 4.06VHH67 pKa = 6.0TPISDD72 pKa = 3.93LPQRR76 pKa = 11.84MLNGFFTQFRR86 pKa = 11.84KK87 pKa = 9.94FKK89 pKa = 10.6YY90 pKa = 10.0AGADD94 pKa = 3.34VTLVPVSTLPADD106 pKa = 3.98PLQISYY112 pKa = 10.45EE113 pKa = 4.07AGEE116 pKa = 4.09PTIDD120 pKa = 3.8PRR122 pKa = 11.84DD123 pKa = 3.72MVNPILWKK131 pKa = 10.26GMSGASMGRR140 pKa = 11.84FLDD143 pKa = 4.76AIFGSHH149 pKa = 7.37RR150 pKa = 11.84IYY152 pKa = 10.7SAQALGDD159 pKa = 3.75VMDD162 pKa = 5.09NSPASEE168 pKa = 3.85TDD170 pKa = 4.25FYY172 pKa = 11.67QLFGDD177 pKa = 4.06SLSISEE183 pKa = 4.29STVFKK188 pKa = 11.07ALTDD192 pKa = 4.05PDD194 pKa = 3.61AVAALEE200 pKa = 3.78AMYY203 pKa = 10.7YY204 pKa = 9.99QGLADD209 pKa = 3.98NSWNKK214 pKa = 10.71AGVQTGFKK222 pKa = 10.3KK223 pKa = 10.49RR224 pKa = 11.84GLFPLVWKK232 pKa = 10.23SASNLFRR239 pKa = 11.84PANQNSPPNPTGTGTPWVPATNVDD263 pKa = 3.51DD264 pKa = 4.17YY265 pKa = 12.07NNYY268 pKa = 10.46GYY270 pKa = 10.68DD271 pKa = 3.53QIHH274 pKa = 6.65EE275 pKa = 4.57SGSPLTPSVGVGIAGGQFSNGTTRR299 pKa = 11.84LGWMPTQTSYY309 pKa = 11.91GMVGTQPSIANAVNGAQDD327 pKa = 3.64MTILPLIPMMMIILPPAYY345 pKa = 8.19KK346 pKa = 9.62TEE348 pKa = 3.83MYY350 pKa = 10.12FRR352 pKa = 11.84MVIKK356 pKa = 10.44HH357 pKa = 5.6KK358 pKa = 11.14FEE360 pKa = 4.13FAEE363 pKa = 4.37FANTVGPDD371 pKa = 3.32TFSNNQNVSPAFIYY385 pKa = 10.78TNVTSGASVATTTEE399 pKa = 4.15EE400 pKa = 3.78QPKK403 pKa = 10.16IVTAGVAA410 pKa = 3.17
MM1 pKa = 7.63GLRR4 pKa = 11.84RR5 pKa = 11.84NIPTAPMLTALWPALLGISLKK26 pKa = 10.61ARR28 pKa = 11.84NIFTDD33 pKa = 4.49FTSDD37 pKa = 3.17NMATNRR43 pKa = 11.84AYY45 pKa = 10.53ASFQEE50 pKa = 4.64IYY52 pKa = 10.64DD53 pKa = 3.36FHH55 pKa = 7.65TEE57 pKa = 3.83VGNTTLIEE65 pKa = 4.06VHH67 pKa = 6.0TPISDD72 pKa = 3.93LPQRR76 pKa = 11.84MLNGFFTQFRR86 pKa = 11.84KK87 pKa = 9.94FKK89 pKa = 10.6YY90 pKa = 10.0AGADD94 pKa = 3.34VTLVPVSTLPADD106 pKa = 3.98PLQISYY112 pKa = 10.45EE113 pKa = 4.07AGEE116 pKa = 4.09PTIDD120 pKa = 3.8PRR122 pKa = 11.84DD123 pKa = 3.72MVNPILWKK131 pKa = 10.26GMSGASMGRR140 pKa = 11.84FLDD143 pKa = 4.76AIFGSHH149 pKa = 7.37RR150 pKa = 11.84IYY152 pKa = 10.7SAQALGDD159 pKa = 3.75VMDD162 pKa = 5.09NSPASEE168 pKa = 3.85TDD170 pKa = 4.25FYY172 pKa = 11.67QLFGDD177 pKa = 4.06SLSISEE183 pKa = 4.29STVFKK188 pKa = 11.07ALTDD192 pKa = 4.05PDD194 pKa = 3.61AVAALEE200 pKa = 3.78AMYY203 pKa = 10.7YY204 pKa = 9.99QGLADD209 pKa = 3.98NSWNKK214 pKa = 10.71AGVQTGFKK222 pKa = 10.3KK223 pKa = 10.49RR224 pKa = 11.84GLFPLVWKK232 pKa = 10.23SASNLFRR239 pKa = 11.84PANQNSPPNPTGTGTPWVPATNVDD263 pKa = 3.51DD264 pKa = 4.17YY265 pKa = 12.07NNYY268 pKa = 10.46GYY270 pKa = 10.68DD271 pKa = 3.53QIHH274 pKa = 6.65EE275 pKa = 4.57SGSPLTPSVGVGIAGGQFSNGTTRR299 pKa = 11.84LGWMPTQTSYY309 pKa = 11.91GMVGTQPSIANAVNGAQDD327 pKa = 3.64MTILPLIPMMMIILPPAYY345 pKa = 8.19KK346 pKa = 9.62TEE348 pKa = 3.83MYY350 pKa = 10.12FRR352 pKa = 11.84MVIKK356 pKa = 10.44HH357 pKa = 5.6KK358 pKa = 11.14FEE360 pKa = 4.13FAEE363 pKa = 4.37FANTVGPDD371 pKa = 3.32TFSNNQNVSPAFIYY385 pKa = 10.78TNVTSGASVATTTEE399 pKa = 4.15EE400 pKa = 3.78QPKK403 pKa = 10.16IVTAGVAA410 pKa = 3.17
Molecular weight: 44.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GP05|A0A0B5GP05_9VIRU Rep protein OS=Howler monkey associated porprismacovirus 1 OX=2170112 GN=rep PE=4 SV=1
MM1 pKa = 7.14STTASRR7 pKa = 11.84QWSTKK12 pKa = 7.34MSQMYY17 pKa = 9.38MLTIPRR23 pKa = 11.84DD24 pKa = 3.61KK25 pKa = 10.75VSKK28 pKa = 10.28RR29 pKa = 11.84EE30 pKa = 3.91LRR32 pKa = 11.84IMLDD36 pKa = 3.67KK37 pKa = 11.05NDD39 pKa = 3.76CKK41 pKa = 10.81KK42 pKa = 10.12WIIGKK47 pKa = 7.57EE48 pKa = 4.07TGKK51 pKa = 10.3NGYY54 pKa = 8.27KK55 pKa = 9.59HH56 pKa = 4.31WQIRR60 pKa = 11.84LEE62 pKa = 4.12TSNEE66 pKa = 3.89EE67 pKa = 3.92FFDD70 pKa = 3.44WCKK73 pKa = 10.98KK74 pKa = 9.96HH75 pKa = 6.73IPTASIRR82 pKa = 11.84KK83 pKa = 9.58AEE85 pKa = 4.12VPKK88 pKa = 9.91WDD90 pKa = 3.73YY91 pKa = 10.08EE92 pKa = 4.29AKK94 pKa = 10.29EE95 pKa = 4.08GQYY98 pKa = 7.49WTSSDD103 pKa = 3.39RR104 pKa = 11.84TDD106 pKa = 3.91NLIQRR111 pKa = 11.84FGEE114 pKa = 3.8FRR116 pKa = 11.84PNQKK120 pKa = 10.02RR121 pKa = 11.84AIQALRR127 pKa = 11.84ATNDD131 pKa = 3.24RR132 pKa = 11.84EE133 pKa = 4.13VLVWYY138 pKa = 10.07DD139 pKa = 3.09EE140 pKa = 4.29GGNVGKK146 pKa = 10.25SWFTGALWEE155 pKa = 4.39RR156 pKa = 11.84GQAYY160 pKa = 7.24VTPPTVSTVQAMVQFVASLYY180 pKa = 10.56CAIEE184 pKa = 4.28SIKK187 pKa = 11.02DD188 pKa = 3.25GLVYY192 pKa = 10.58DD193 pKa = 3.63SRR195 pKa = 11.84YY196 pKa = 8.17QGRR199 pKa = 11.84MVNIRR204 pKa = 11.84GVKK207 pKa = 9.51IIVMTNNKK215 pKa = 9.0PDD217 pKa = 4.21LDD219 pKa = 3.88KK220 pKa = 11.35LSYY223 pKa = 10.57DD224 pKa = 3.15RR225 pKa = 11.84WRR227 pKa = 11.84MVVLL231 pKa = 4.26
MM1 pKa = 7.14STTASRR7 pKa = 11.84QWSTKK12 pKa = 7.34MSQMYY17 pKa = 9.38MLTIPRR23 pKa = 11.84DD24 pKa = 3.61KK25 pKa = 10.75VSKK28 pKa = 10.28RR29 pKa = 11.84EE30 pKa = 3.91LRR32 pKa = 11.84IMLDD36 pKa = 3.67KK37 pKa = 11.05NDD39 pKa = 3.76CKK41 pKa = 10.81KK42 pKa = 10.12WIIGKK47 pKa = 7.57EE48 pKa = 4.07TGKK51 pKa = 10.3NGYY54 pKa = 8.27KK55 pKa = 9.59HH56 pKa = 4.31WQIRR60 pKa = 11.84LEE62 pKa = 4.12TSNEE66 pKa = 3.89EE67 pKa = 3.92FFDD70 pKa = 3.44WCKK73 pKa = 10.98KK74 pKa = 9.96HH75 pKa = 6.73IPTASIRR82 pKa = 11.84KK83 pKa = 9.58AEE85 pKa = 4.12VPKK88 pKa = 9.91WDD90 pKa = 3.73YY91 pKa = 10.08EE92 pKa = 4.29AKK94 pKa = 10.29EE95 pKa = 4.08GQYY98 pKa = 7.49WTSSDD103 pKa = 3.39RR104 pKa = 11.84TDD106 pKa = 3.91NLIQRR111 pKa = 11.84FGEE114 pKa = 3.8FRR116 pKa = 11.84PNQKK120 pKa = 10.02RR121 pKa = 11.84AIQALRR127 pKa = 11.84ATNDD131 pKa = 3.24RR132 pKa = 11.84EE133 pKa = 4.13VLVWYY138 pKa = 10.07DD139 pKa = 3.09EE140 pKa = 4.29GGNVGKK146 pKa = 10.25SWFTGALWEE155 pKa = 4.39RR156 pKa = 11.84GQAYY160 pKa = 7.24VTPPTVSTVQAMVQFVASLYY180 pKa = 10.56CAIEE184 pKa = 4.28SIKK187 pKa = 11.02DD188 pKa = 3.25GLVYY192 pKa = 10.58DD193 pKa = 3.63SRR195 pKa = 11.84YY196 pKa = 8.17QGRR199 pKa = 11.84MVNIRR204 pKa = 11.84GVKK207 pKa = 9.51IIVMTNNKK215 pKa = 9.0PDD217 pKa = 4.21LDD219 pKa = 3.88KK220 pKa = 11.35LSYY223 pKa = 10.57DD224 pKa = 3.15RR225 pKa = 11.84WRR227 pKa = 11.84MVVLL231 pKa = 4.26
Molecular weight: 27.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
641 |
231 |
410 |
320.5 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.8 ± 1.606 | 0.468 ± 0.512 |
5.46 ± 0.37 | 4.212 ± 0.872 |
4.524 ± 1.187 | 7.02 ± 0.858 |
1.092 ± 0.139 | 5.616 ± 0.274 |
5.148 ± 2.163 | 6.396 ± 0.474 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.056 ± 0.099 | 5.46 ± 0.697 |
5.928 ± 1.786 | 4.212 ± 0.339 |
4.836 ± 1.821 | 7.02 ± 0.325 |
8.112 ± 0.998 | 6.24 ± 0.423 |
2.496 ± 1.129 | 3.9 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
