
Pacific flying fox faeces associated circular DNA virus-9
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
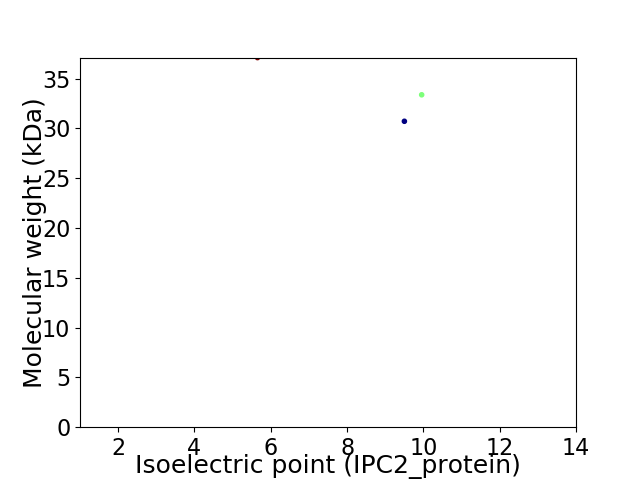
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTJ0|A0A140CTJ0_9VIRU Uncharacterized protein OS=Pacific flying fox faeces associated circular DNA virus-9 OX=1796018 PE=4 SV=1
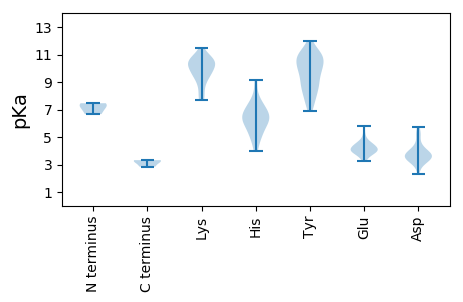
MM1 pKa = 7.27FAKK4 pKa = 10.43FWLITAHH11 pKa = 6.69FDD13 pKa = 3.57EE14 pKa = 5.83EE15 pKa = 4.47EE16 pKa = 3.98FLDD19 pKa = 5.23LVTTEE24 pKa = 4.71LGEE27 pKa = 4.06LSPVIRR33 pKa = 11.84FVWWQLEE40 pKa = 3.96AGEE43 pKa = 4.39QRR45 pKa = 11.84GSLHH49 pKa = 4.86VQAYY53 pKa = 9.93LGLHH57 pKa = 6.1RR58 pKa = 11.84SQRR61 pKa = 11.84LSYY64 pKa = 10.95VVGLFDD70 pKa = 3.72GASRR74 pKa = 11.84PVHH77 pKa = 6.0AEE79 pKa = 3.52PRR81 pKa = 11.84RR82 pKa = 11.84GTHH85 pKa = 5.13EE86 pKa = 3.94QAIQYY91 pKa = 7.4CTKK94 pKa = 10.24EE95 pKa = 4.0DD96 pKa = 3.38TRR98 pKa = 11.84RR99 pKa = 11.84AGPWFLGQPEE109 pKa = 4.35EE110 pKa = 4.29TGPGQSNALLDD121 pKa = 3.67IRR123 pKa = 11.84DD124 pKa = 3.84GLDD127 pKa = 2.96RR128 pKa = 11.84GVTLVEE134 pKa = 5.17LARR137 pKa = 11.84QDD139 pKa = 3.93NLFGALIRR147 pKa = 11.84NIRR150 pKa = 11.84GLQWYY155 pKa = 8.07AAQTGVDD162 pKa = 4.16RR163 pKa = 11.84NWEE166 pKa = 4.06TTVIILSGAPGVGKK180 pKa = 10.47SRR182 pKa = 11.84AAEE185 pKa = 3.66AAARR189 pKa = 11.84IVGVPYY195 pKa = 10.87YY196 pKa = 9.45LTRR199 pKa = 11.84PQAANSLPWWDD210 pKa = 4.48GYY212 pKa = 9.63TPGAPVVIDD221 pKa = 5.74DD222 pKa = 4.36FYY224 pKa = 11.95GWIKK228 pKa = 10.6LDD230 pKa = 3.33TMLRR234 pKa = 11.84LMDD237 pKa = 5.03RR238 pKa = 11.84YY239 pKa = 10.14PMQVEE244 pKa = 4.68TKK246 pKa = 10.63GGMLKK251 pKa = 9.29FTSRR255 pKa = 11.84LMIVTSNYY263 pKa = 7.27EE264 pKa = 3.27WRR266 pKa = 11.84NWYY269 pKa = 8.02RR270 pKa = 11.84TAGNAHH276 pKa = 6.76FPRR279 pKa = 11.84NFEE282 pKa = 3.74RR283 pKa = 11.84RR284 pKa = 11.84INRR287 pKa = 11.84DD288 pKa = 2.29IYY290 pKa = 8.79MTAEE294 pKa = 4.48DD295 pKa = 5.3DD296 pKa = 3.73YY297 pKa = 11.68LDD299 pKa = 3.36IVRR302 pKa = 11.84SILQVYY308 pKa = 10.62NDD310 pKa = 3.64MHH312 pKa = 6.23NTFHH316 pKa = 7.34WICDD320 pKa = 3.82YY321 pKa = 11.4VV322 pKa = 3.32
MM1 pKa = 7.27FAKK4 pKa = 10.43FWLITAHH11 pKa = 6.69FDD13 pKa = 3.57EE14 pKa = 5.83EE15 pKa = 4.47EE16 pKa = 3.98FLDD19 pKa = 5.23LVTTEE24 pKa = 4.71LGEE27 pKa = 4.06LSPVIRR33 pKa = 11.84FVWWQLEE40 pKa = 3.96AGEE43 pKa = 4.39QRR45 pKa = 11.84GSLHH49 pKa = 4.86VQAYY53 pKa = 9.93LGLHH57 pKa = 6.1RR58 pKa = 11.84SQRR61 pKa = 11.84LSYY64 pKa = 10.95VVGLFDD70 pKa = 3.72GASRR74 pKa = 11.84PVHH77 pKa = 6.0AEE79 pKa = 3.52PRR81 pKa = 11.84RR82 pKa = 11.84GTHH85 pKa = 5.13EE86 pKa = 3.94QAIQYY91 pKa = 7.4CTKK94 pKa = 10.24EE95 pKa = 4.0DD96 pKa = 3.38TRR98 pKa = 11.84RR99 pKa = 11.84AGPWFLGQPEE109 pKa = 4.35EE110 pKa = 4.29TGPGQSNALLDD121 pKa = 3.67IRR123 pKa = 11.84DD124 pKa = 3.84GLDD127 pKa = 2.96RR128 pKa = 11.84GVTLVEE134 pKa = 5.17LARR137 pKa = 11.84QDD139 pKa = 3.93NLFGALIRR147 pKa = 11.84NIRR150 pKa = 11.84GLQWYY155 pKa = 8.07AAQTGVDD162 pKa = 4.16RR163 pKa = 11.84NWEE166 pKa = 4.06TTVIILSGAPGVGKK180 pKa = 10.47SRR182 pKa = 11.84AAEE185 pKa = 3.66AAARR189 pKa = 11.84IVGVPYY195 pKa = 10.87YY196 pKa = 9.45LTRR199 pKa = 11.84PQAANSLPWWDD210 pKa = 4.48GYY212 pKa = 9.63TPGAPVVIDD221 pKa = 5.74DD222 pKa = 4.36FYY224 pKa = 11.95GWIKK228 pKa = 10.6LDD230 pKa = 3.33TMLRR234 pKa = 11.84LMDD237 pKa = 5.03RR238 pKa = 11.84YY239 pKa = 10.14PMQVEE244 pKa = 4.68TKK246 pKa = 10.63GGMLKK251 pKa = 9.29FTSRR255 pKa = 11.84LMIVTSNYY263 pKa = 7.27EE264 pKa = 3.27WRR266 pKa = 11.84NWYY269 pKa = 8.02RR270 pKa = 11.84TAGNAHH276 pKa = 6.76FPRR279 pKa = 11.84NFEE282 pKa = 3.74RR283 pKa = 11.84RR284 pKa = 11.84INRR287 pKa = 11.84DD288 pKa = 2.29IYY290 pKa = 8.79MTAEE294 pKa = 4.48DD295 pKa = 5.3DD296 pKa = 3.73YY297 pKa = 11.68LDD299 pKa = 3.36IVRR302 pKa = 11.84SILQVYY308 pKa = 10.62NDD310 pKa = 3.64MHH312 pKa = 6.23NTFHH316 pKa = 7.34WICDD320 pKa = 3.82YY321 pKa = 11.4VV322 pKa = 3.32
Molecular weight: 37.11 kDa
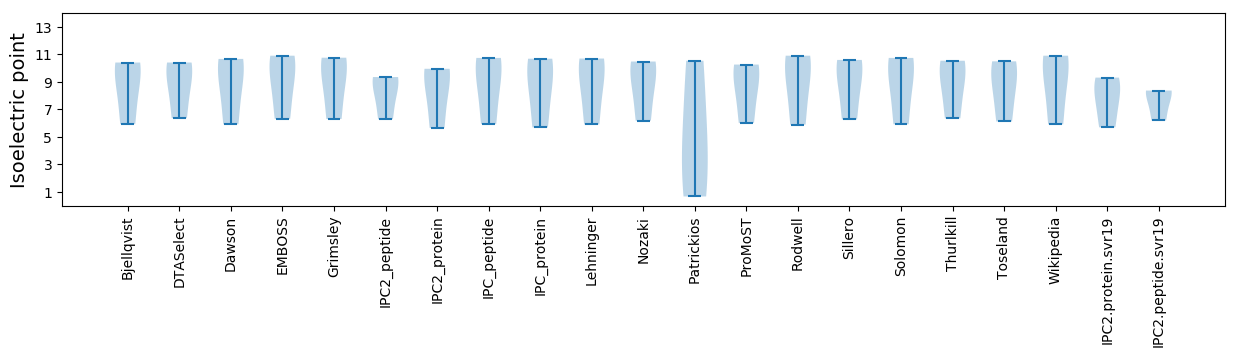
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTJ1|A0A140CTJ1_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-9 OX=1796018 PE=4 SV=1
MM1 pKa = 6.67TAYY4 pKa = 10.18RR5 pKa = 11.84QQRR8 pKa = 11.84QQQYY12 pKa = 9.45RR13 pKa = 11.84QQQYY17 pKa = 8.85LQYY20 pKa = 8.45RR21 pKa = 11.84QFWRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 7.36SRR29 pKa = 11.84NMPAKK34 pKa = 9.75RR35 pKa = 11.84VRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84KK41 pKa = 7.69STYY44 pKa = 6.87TRR46 pKa = 11.84RR47 pKa = 11.84ARR49 pKa = 11.84ARR51 pKa = 11.84RR52 pKa = 11.84TDD54 pKa = 2.83MRR56 pKa = 11.84TQRR59 pKa = 11.84MIVNSFYY66 pKa = 11.11GRR68 pKa = 11.84SEE70 pKa = 4.08TKK72 pKa = 10.4LLQVRR77 pKa = 11.84SGAYY81 pKa = 9.39LVNGAVGDD89 pKa = 3.89GLRR92 pKa = 11.84VDD94 pKa = 3.86SSLVQPVYY102 pKa = 11.15GKK104 pKa = 8.42ITQGSAGDD112 pKa = 3.61QRR114 pKa = 11.84IGNQIFVKK122 pKa = 9.94TISICFTIANNSFLTVDD139 pKa = 3.22QPMINGQTNVGYY151 pKa = 8.9PIPVKK156 pKa = 10.91NNIAGRR162 pKa = 11.84PDD164 pKa = 3.31RR165 pKa = 11.84GMVLQPKK172 pKa = 9.62DD173 pKa = 3.33SKK175 pKa = 11.45YY176 pKa = 10.6DD177 pKa = 3.49QEE179 pKa = 4.5GSKK182 pKa = 10.31FLPYY186 pKa = 9.68LTGFLAGTAMNAQHH200 pKa = 7.33DD201 pKa = 4.25LRR203 pKa = 11.84YY204 pKa = 10.81VNVLYY209 pKa = 10.9DD210 pKa = 3.75RR211 pKa = 11.84MEE213 pKa = 4.21VMPSTWSRR221 pKa = 11.84NNSIVGSRR229 pKa = 11.84STKK232 pKa = 9.64IVRR235 pKa = 11.84KK236 pKa = 9.41INKK239 pKa = 8.78KK240 pKa = 7.91VQYY243 pKa = 8.85EE244 pKa = 4.06QGAGTFVKK252 pKa = 10.42GGEE255 pKa = 4.23VFLCIMPGFSVADD268 pKa = 3.82PSTAVTGADD277 pKa = 3.28NKK279 pKa = 10.77PSFFSQMYY287 pKa = 10.45VSFTDD292 pKa = 3.3SS293 pKa = 2.84
MM1 pKa = 6.67TAYY4 pKa = 10.18RR5 pKa = 11.84QQRR8 pKa = 11.84QQQYY12 pKa = 9.45RR13 pKa = 11.84QQQYY17 pKa = 8.85LQYY20 pKa = 8.45RR21 pKa = 11.84QFWRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 7.36SRR29 pKa = 11.84NMPAKK34 pKa = 9.75RR35 pKa = 11.84VRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84KK41 pKa = 7.69STYY44 pKa = 6.87TRR46 pKa = 11.84RR47 pKa = 11.84ARR49 pKa = 11.84ARR51 pKa = 11.84RR52 pKa = 11.84TDD54 pKa = 2.83MRR56 pKa = 11.84TQRR59 pKa = 11.84MIVNSFYY66 pKa = 11.11GRR68 pKa = 11.84SEE70 pKa = 4.08TKK72 pKa = 10.4LLQVRR77 pKa = 11.84SGAYY81 pKa = 9.39LVNGAVGDD89 pKa = 3.89GLRR92 pKa = 11.84VDD94 pKa = 3.86SSLVQPVYY102 pKa = 11.15GKK104 pKa = 8.42ITQGSAGDD112 pKa = 3.61QRR114 pKa = 11.84IGNQIFVKK122 pKa = 9.94TISICFTIANNSFLTVDD139 pKa = 3.22QPMINGQTNVGYY151 pKa = 8.9PIPVKK156 pKa = 10.91NNIAGRR162 pKa = 11.84PDD164 pKa = 3.31RR165 pKa = 11.84GMVLQPKK172 pKa = 9.62DD173 pKa = 3.33SKK175 pKa = 11.45YY176 pKa = 10.6DD177 pKa = 3.49QEE179 pKa = 4.5GSKK182 pKa = 10.31FLPYY186 pKa = 9.68LTGFLAGTAMNAQHH200 pKa = 7.33DD201 pKa = 4.25LRR203 pKa = 11.84YY204 pKa = 10.81VNVLYY209 pKa = 10.9DD210 pKa = 3.75RR211 pKa = 11.84MEE213 pKa = 4.21VMPSTWSRR221 pKa = 11.84NNSIVGSRR229 pKa = 11.84STKK232 pKa = 9.64IVRR235 pKa = 11.84KK236 pKa = 9.41INKK239 pKa = 8.78KK240 pKa = 7.91VQYY243 pKa = 8.85EE244 pKa = 4.06QGAGTFVKK252 pKa = 10.42GGEE255 pKa = 4.23VFLCIMPGFSVADD268 pKa = 3.82PSTAVTGADD277 pKa = 3.28NKK279 pKa = 10.77PSFFSQMYY287 pKa = 10.45VSFTDD292 pKa = 3.3SS293 pKa = 2.84
Molecular weight: 33.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
876 |
261 |
322 |
292.0 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.708 ± 1.147 | 2.169 ± 1.508 |
3.881 ± 1.577 | 2.968 ± 1.423 |
4.452 ± 0.251 | 6.279 ± 1.682 |
2.283 ± 0.897 | 6.621 ± 1.557 |
3.881 ± 0.932 | 7.306 ± 1.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.511 ± 0.61 | 4.224 ± 0.484 |
4.566 ± 0.178 | 5.594 ± 0.998 |
8.447 ± 0.826 | 6.735 ± 1.493 |
7.42 ± 1.079 | 6.621 ± 0.747 |
1.941 ± 0.816 | 6.393 ± 1.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
