
Cronartium ribicola mitovirus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191KCP1|A0A191KCP1_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 4 OX=1816487 GN=RdRp PE=4 SV=1
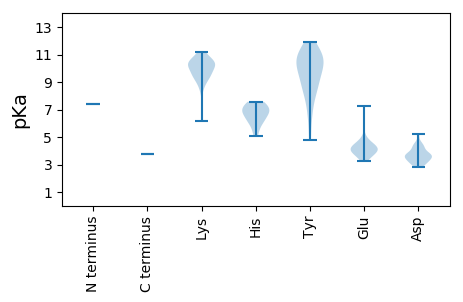
MM1 pKa = 7.44KK2 pKa = 10.63LIINIMKK9 pKa = 9.69IWAQSKK15 pKa = 10.17RR16 pKa = 11.84FLQIAKK22 pKa = 8.67TFYY25 pKa = 8.4TWYY28 pKa = 9.68SVYY31 pKa = 10.77YY32 pKa = 6.13PTCSPDD38 pKa = 4.25DD39 pKa = 3.21IAPFLEE45 pKa = 4.5RR46 pKa = 11.84VHH48 pKa = 6.88HH49 pKa = 6.03SLANRR54 pKa = 11.84GALATLEE61 pKa = 4.13WLKK64 pKa = 9.19EE65 pKa = 3.82TRR67 pKa = 11.84LAFTRR72 pKa = 11.84WICGDD77 pKa = 3.67SRR79 pKa = 11.84PDD81 pKa = 3.17VCVKK85 pKa = 10.42LDD87 pKa = 3.68SSGLPVMVKK96 pKa = 8.84STLGKK101 pKa = 10.5FIDD104 pKa = 3.93TARR107 pKa = 11.84GRR109 pKa = 11.84ALILTLLNCIRR120 pKa = 11.84SYY122 pKa = 11.33EE123 pKa = 4.09GLKK126 pKa = 10.37EE127 pKa = 3.93PSISDD132 pKa = 2.85ITSPYY137 pKa = 10.03KK138 pKa = 9.87GEE140 pKa = 3.84KK141 pKa = 9.38WIDD144 pKa = 3.52SYY146 pKa = 11.9LGLYY150 pKa = 8.75TKK152 pKa = 10.81DD153 pKa = 3.64FLRR156 pKa = 11.84KK157 pKa = 9.59YY158 pKa = 10.92GDD160 pKa = 2.92IHH162 pKa = 7.43RR163 pKa = 11.84PRR165 pKa = 11.84PQFKK169 pKa = 9.21QWLYY173 pKa = 4.77TTKK176 pKa = 10.37RR177 pKa = 11.84GPNGPSIATAYY188 pKa = 10.65HH189 pKa = 7.57DD190 pKa = 3.29ITLLTEE196 pKa = 3.45EE197 pKa = 5.03HH198 pKa = 5.69YY199 pKa = 10.7RR200 pKa = 11.84IYY202 pKa = 10.83EE203 pKa = 3.98VLAGKK208 pKa = 9.8DD209 pKa = 3.72YY210 pKa = 11.16KK211 pKa = 11.18DD212 pKa = 3.42SLLKK216 pKa = 10.4IKK218 pKa = 10.26EE219 pKa = 4.18IASDD223 pKa = 3.5PQEE226 pKa = 3.95FEE228 pKa = 4.9RR229 pKa = 11.84IRR231 pKa = 11.84KK232 pKa = 9.23DD233 pKa = 3.08FSLIVDD239 pKa = 3.8IKK241 pKa = 10.57SRR243 pKa = 11.84YY244 pKa = 6.84VEE246 pKa = 4.58KK247 pKa = 10.63ISVIPSPEE255 pKa = 3.51GKK257 pKa = 8.79MRR259 pKa = 11.84TISIASYY266 pKa = 8.65WAQNALDD273 pKa = 4.33PLHH276 pKa = 6.91RR277 pKa = 11.84CIFKK281 pKa = 10.37YY282 pKa = 10.27LRR284 pKa = 11.84KK285 pKa = 9.54IPNDD289 pKa = 2.89ITFDD293 pKa = 3.52QSKK296 pKa = 10.32GLSILNKK303 pKa = 9.66GQNDD307 pKa = 4.53SYY309 pKa = 11.11WSYY312 pKa = 11.6DD313 pKa = 3.23LKK315 pKa = 11.16AATDD319 pKa = 3.88RR320 pKa = 11.84FPRR323 pKa = 11.84STQTLLLSHH332 pKa = 7.51LIGQEE337 pKa = 4.35FANTWSLLISAEE349 pKa = 3.84HH350 pKa = 7.51DD351 pKa = 3.11IPAIGIRR358 pKa = 11.84AAWAVGQPIGKK369 pKa = 9.2LSSWAVFAYY378 pKa = 7.38THH380 pKa = 6.33HH381 pKa = 6.61FIIYY385 pKa = 7.55VAHH388 pKa = 6.99RR389 pKa = 11.84RR390 pKa = 11.84ACVPFAPGTYY400 pKa = 10.06VILGDD405 pKa = 4.33DD406 pKa = 3.66VVIANDD412 pKa = 3.75KK413 pKa = 10.43VAEE416 pKa = 4.16QYY418 pKa = 8.64STLIRR423 pKa = 11.84WLGVEE428 pKa = 3.41ISMNKK433 pKa = 6.13THH435 pKa = 7.2RR436 pKa = 11.84SSHH439 pKa = 5.05SFEE442 pKa = 3.73IGKK445 pKa = 9.53RR446 pKa = 11.84WIHH449 pKa = 6.41RR450 pKa = 11.84GQEE453 pKa = 3.6ISPFSIVPWMQAKK466 pKa = 9.37NWAPAITQCIYY477 pKa = 10.66EE478 pKa = 4.19EE479 pKa = 4.63KK480 pKa = 10.04KK481 pKa = 10.81RR482 pKa = 11.84SWSLPFHH489 pKa = 6.12PVEE492 pKa = 4.38GVARR496 pKa = 11.84CILLQYY502 pKa = 9.86HH503 pKa = 6.86GKK505 pKa = 10.31LSSRR509 pKa = 11.84YY510 pKa = 8.54IDD512 pKa = 3.6HH513 pKa = 7.28LFDD516 pKa = 5.22LSQLNVAVYY525 pKa = 9.84HH526 pKa = 6.35SIKK529 pKa = 10.83EE530 pKa = 4.03MITWSDD536 pKa = 4.16CINIIAEE543 pKa = 4.24VFYY546 pKa = 9.4KK547 pKa = 9.79TKK549 pKa = 10.0WEE551 pKa = 4.12HH552 pKa = 6.75LDD554 pKa = 3.49RR555 pKa = 11.84LHH557 pKa = 6.92GRR559 pKa = 11.84LVQTYY564 pKa = 9.57VYY566 pKa = 10.86LRR568 pKa = 11.84LFNKK572 pKa = 9.88NAIPHH577 pKa = 5.98WSAIKK582 pKa = 10.36KK583 pKa = 10.14VILVEE588 pKa = 4.34NGFSMASIIQIIMGGRR604 pKa = 11.84SLSGASFSYY613 pKa = 11.39ADD615 pKa = 4.58LPHH618 pKa = 6.57KK619 pKa = 10.54KK620 pKa = 9.7IIRR623 pKa = 11.84RR624 pKa = 11.84YY625 pKa = 8.3AQEE628 pKa = 4.55FFDD631 pKa = 4.31KK632 pKa = 11.12NADD635 pKa = 3.61VVQGHH640 pKa = 5.4YY641 pKa = 10.68FEE643 pKa = 7.28DD644 pKa = 3.84GMLQRR649 pKa = 11.84EE650 pKa = 4.6AVRR653 pKa = 11.84SLGIPNVEE661 pKa = 4.02HH662 pKa = 7.27AFTKK666 pKa = 10.17SKK668 pKa = 10.22VRR670 pKa = 11.84RR671 pKa = 11.84EE672 pKa = 4.21VILSNNILKK681 pKa = 9.37EE682 pKa = 3.7LCSFVKK688 pKa = 10.36SVPSSGFTHH697 pKa = 6.99TSGSPVPTEE706 pKa = 3.26INEE709 pKa = 4.34YY710 pKa = 8.9IRR712 pKa = 11.84KK713 pKa = 8.31VNYY716 pKa = 9.8YY717 pKa = 10.05EE718 pKa = 4.01WDD720 pKa = 3.3TT721 pKa = 3.76
MM1 pKa = 7.44KK2 pKa = 10.63LIINIMKK9 pKa = 9.69IWAQSKK15 pKa = 10.17RR16 pKa = 11.84FLQIAKK22 pKa = 8.67TFYY25 pKa = 8.4TWYY28 pKa = 9.68SVYY31 pKa = 10.77YY32 pKa = 6.13PTCSPDD38 pKa = 4.25DD39 pKa = 3.21IAPFLEE45 pKa = 4.5RR46 pKa = 11.84VHH48 pKa = 6.88HH49 pKa = 6.03SLANRR54 pKa = 11.84GALATLEE61 pKa = 4.13WLKK64 pKa = 9.19EE65 pKa = 3.82TRR67 pKa = 11.84LAFTRR72 pKa = 11.84WICGDD77 pKa = 3.67SRR79 pKa = 11.84PDD81 pKa = 3.17VCVKK85 pKa = 10.42LDD87 pKa = 3.68SSGLPVMVKK96 pKa = 8.84STLGKK101 pKa = 10.5FIDD104 pKa = 3.93TARR107 pKa = 11.84GRR109 pKa = 11.84ALILTLLNCIRR120 pKa = 11.84SYY122 pKa = 11.33EE123 pKa = 4.09GLKK126 pKa = 10.37EE127 pKa = 3.93PSISDD132 pKa = 2.85ITSPYY137 pKa = 10.03KK138 pKa = 9.87GEE140 pKa = 3.84KK141 pKa = 9.38WIDD144 pKa = 3.52SYY146 pKa = 11.9LGLYY150 pKa = 8.75TKK152 pKa = 10.81DD153 pKa = 3.64FLRR156 pKa = 11.84KK157 pKa = 9.59YY158 pKa = 10.92GDD160 pKa = 2.92IHH162 pKa = 7.43RR163 pKa = 11.84PRR165 pKa = 11.84PQFKK169 pKa = 9.21QWLYY173 pKa = 4.77TTKK176 pKa = 10.37RR177 pKa = 11.84GPNGPSIATAYY188 pKa = 10.65HH189 pKa = 7.57DD190 pKa = 3.29ITLLTEE196 pKa = 3.45EE197 pKa = 5.03HH198 pKa = 5.69YY199 pKa = 10.7RR200 pKa = 11.84IYY202 pKa = 10.83EE203 pKa = 3.98VLAGKK208 pKa = 9.8DD209 pKa = 3.72YY210 pKa = 11.16KK211 pKa = 11.18DD212 pKa = 3.42SLLKK216 pKa = 10.4IKK218 pKa = 10.26EE219 pKa = 4.18IASDD223 pKa = 3.5PQEE226 pKa = 3.95FEE228 pKa = 4.9RR229 pKa = 11.84IRR231 pKa = 11.84KK232 pKa = 9.23DD233 pKa = 3.08FSLIVDD239 pKa = 3.8IKK241 pKa = 10.57SRR243 pKa = 11.84YY244 pKa = 6.84VEE246 pKa = 4.58KK247 pKa = 10.63ISVIPSPEE255 pKa = 3.51GKK257 pKa = 8.79MRR259 pKa = 11.84TISIASYY266 pKa = 8.65WAQNALDD273 pKa = 4.33PLHH276 pKa = 6.91RR277 pKa = 11.84CIFKK281 pKa = 10.37YY282 pKa = 10.27LRR284 pKa = 11.84KK285 pKa = 9.54IPNDD289 pKa = 2.89ITFDD293 pKa = 3.52QSKK296 pKa = 10.32GLSILNKK303 pKa = 9.66GQNDD307 pKa = 4.53SYY309 pKa = 11.11WSYY312 pKa = 11.6DD313 pKa = 3.23LKK315 pKa = 11.16AATDD319 pKa = 3.88RR320 pKa = 11.84FPRR323 pKa = 11.84STQTLLLSHH332 pKa = 7.51LIGQEE337 pKa = 4.35FANTWSLLISAEE349 pKa = 3.84HH350 pKa = 7.51DD351 pKa = 3.11IPAIGIRR358 pKa = 11.84AAWAVGQPIGKK369 pKa = 9.2LSSWAVFAYY378 pKa = 7.38THH380 pKa = 6.33HH381 pKa = 6.61FIIYY385 pKa = 7.55VAHH388 pKa = 6.99RR389 pKa = 11.84RR390 pKa = 11.84ACVPFAPGTYY400 pKa = 10.06VILGDD405 pKa = 4.33DD406 pKa = 3.66VVIANDD412 pKa = 3.75KK413 pKa = 10.43VAEE416 pKa = 4.16QYY418 pKa = 8.64STLIRR423 pKa = 11.84WLGVEE428 pKa = 3.41ISMNKK433 pKa = 6.13THH435 pKa = 7.2RR436 pKa = 11.84SSHH439 pKa = 5.05SFEE442 pKa = 3.73IGKK445 pKa = 9.53RR446 pKa = 11.84WIHH449 pKa = 6.41RR450 pKa = 11.84GQEE453 pKa = 3.6ISPFSIVPWMQAKK466 pKa = 9.37NWAPAITQCIYY477 pKa = 10.66EE478 pKa = 4.19EE479 pKa = 4.63KK480 pKa = 10.04KK481 pKa = 10.81RR482 pKa = 11.84SWSLPFHH489 pKa = 6.12PVEE492 pKa = 4.38GVARR496 pKa = 11.84CILLQYY502 pKa = 9.86HH503 pKa = 6.86GKK505 pKa = 10.31LSSRR509 pKa = 11.84YY510 pKa = 8.54IDD512 pKa = 3.6HH513 pKa = 7.28LFDD516 pKa = 5.22LSQLNVAVYY525 pKa = 9.84HH526 pKa = 6.35SIKK529 pKa = 10.83EE530 pKa = 4.03MITWSDD536 pKa = 4.16CINIIAEE543 pKa = 4.24VFYY546 pKa = 9.4KK547 pKa = 9.79TKK549 pKa = 10.0WEE551 pKa = 4.12HH552 pKa = 6.75LDD554 pKa = 3.49RR555 pKa = 11.84LHH557 pKa = 6.92GRR559 pKa = 11.84LVQTYY564 pKa = 9.57VYY566 pKa = 10.86LRR568 pKa = 11.84LFNKK572 pKa = 9.88NAIPHH577 pKa = 5.98WSAIKK582 pKa = 10.36KK583 pKa = 10.14VILVEE588 pKa = 4.34NGFSMASIIQIIMGGRR604 pKa = 11.84SLSGASFSYY613 pKa = 11.39ADD615 pKa = 4.58LPHH618 pKa = 6.57KK619 pKa = 10.54KK620 pKa = 9.7IIRR623 pKa = 11.84RR624 pKa = 11.84YY625 pKa = 8.3AQEE628 pKa = 4.55FFDD631 pKa = 4.31KK632 pKa = 11.12NADD635 pKa = 3.61VVQGHH640 pKa = 5.4YY641 pKa = 10.68FEE643 pKa = 7.28DD644 pKa = 3.84GMLQRR649 pKa = 11.84EE650 pKa = 4.6AVRR653 pKa = 11.84SLGIPNVEE661 pKa = 4.02HH662 pKa = 7.27AFTKK666 pKa = 10.17SKK668 pKa = 10.22VRR670 pKa = 11.84RR671 pKa = 11.84EE672 pKa = 4.21VILSNNILKK681 pKa = 9.37EE682 pKa = 3.7LCSFVKK688 pKa = 10.36SVPSSGFTHH697 pKa = 6.99TSGSPVPTEE706 pKa = 3.26INEE709 pKa = 4.34YY710 pKa = 8.9IRR712 pKa = 11.84KK713 pKa = 8.31VNYY716 pKa = 9.8YY717 pKa = 10.05EE718 pKa = 4.01WDD720 pKa = 3.3TT721 pKa = 3.76
Molecular weight: 83.11 kDa
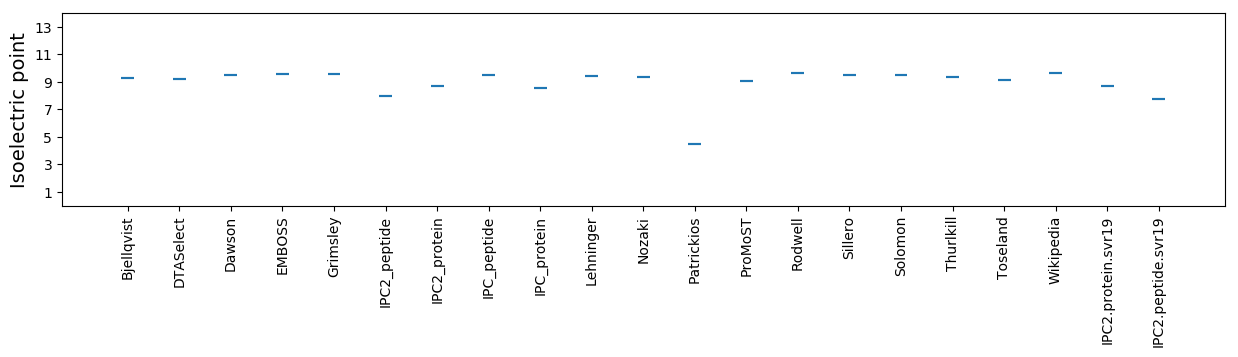
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191KCP1|A0A191KCP1_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 4 OX=1816487 GN=RdRp PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.63LIINIMKK9 pKa = 9.69IWAQSKK15 pKa = 10.17RR16 pKa = 11.84FLQIAKK22 pKa = 8.67TFYY25 pKa = 8.4TWYY28 pKa = 9.68SVYY31 pKa = 10.77YY32 pKa = 6.13PTCSPDD38 pKa = 4.25DD39 pKa = 3.21IAPFLEE45 pKa = 4.5RR46 pKa = 11.84VHH48 pKa = 6.88HH49 pKa = 6.03SLANRR54 pKa = 11.84GALATLEE61 pKa = 4.13WLKK64 pKa = 9.19EE65 pKa = 3.82TRR67 pKa = 11.84LAFTRR72 pKa = 11.84WICGDD77 pKa = 3.67SRR79 pKa = 11.84PDD81 pKa = 3.17VCVKK85 pKa = 10.42LDD87 pKa = 3.68SSGLPVMVKK96 pKa = 8.84STLGKK101 pKa = 10.5FIDD104 pKa = 3.93TARR107 pKa = 11.84GRR109 pKa = 11.84ALILTLLNCIRR120 pKa = 11.84SYY122 pKa = 11.33EE123 pKa = 4.09GLKK126 pKa = 10.37EE127 pKa = 3.93PSISDD132 pKa = 2.85ITSPYY137 pKa = 10.03KK138 pKa = 9.87GEE140 pKa = 3.84KK141 pKa = 9.38WIDD144 pKa = 3.52SYY146 pKa = 11.9LGLYY150 pKa = 8.75TKK152 pKa = 10.81DD153 pKa = 3.64FLRR156 pKa = 11.84KK157 pKa = 9.59YY158 pKa = 10.92GDD160 pKa = 2.92IHH162 pKa = 7.43RR163 pKa = 11.84PRR165 pKa = 11.84PQFKK169 pKa = 9.21QWLYY173 pKa = 4.77TTKK176 pKa = 10.37RR177 pKa = 11.84GPNGPSIATAYY188 pKa = 10.65HH189 pKa = 7.57DD190 pKa = 3.29ITLLTEE196 pKa = 3.45EE197 pKa = 5.03HH198 pKa = 5.69YY199 pKa = 10.7RR200 pKa = 11.84IYY202 pKa = 10.83EE203 pKa = 3.98VLAGKK208 pKa = 9.8DD209 pKa = 3.72YY210 pKa = 11.16KK211 pKa = 11.18DD212 pKa = 3.42SLLKK216 pKa = 10.4IKK218 pKa = 10.26EE219 pKa = 4.18IASDD223 pKa = 3.5PQEE226 pKa = 3.95FEE228 pKa = 4.9RR229 pKa = 11.84IRR231 pKa = 11.84KK232 pKa = 9.23DD233 pKa = 3.08FSLIVDD239 pKa = 3.8IKK241 pKa = 10.57SRR243 pKa = 11.84YY244 pKa = 6.84VEE246 pKa = 4.58KK247 pKa = 10.63ISVIPSPEE255 pKa = 3.51GKK257 pKa = 8.79MRR259 pKa = 11.84TISIASYY266 pKa = 8.65WAQNALDD273 pKa = 4.33PLHH276 pKa = 6.91RR277 pKa = 11.84CIFKK281 pKa = 10.37YY282 pKa = 10.27LRR284 pKa = 11.84KK285 pKa = 9.54IPNDD289 pKa = 2.89ITFDD293 pKa = 3.52QSKK296 pKa = 10.32GLSILNKK303 pKa = 9.66GQNDD307 pKa = 4.53SYY309 pKa = 11.11WSYY312 pKa = 11.6DD313 pKa = 3.23LKK315 pKa = 11.16AATDD319 pKa = 3.88RR320 pKa = 11.84FPRR323 pKa = 11.84STQTLLLSHH332 pKa = 7.51LIGQEE337 pKa = 4.35FANTWSLLISAEE349 pKa = 3.84HH350 pKa = 7.51DD351 pKa = 3.11IPAIGIRR358 pKa = 11.84AAWAVGQPIGKK369 pKa = 9.2LSSWAVFAYY378 pKa = 7.38THH380 pKa = 6.33HH381 pKa = 6.61FIIYY385 pKa = 7.55VAHH388 pKa = 6.99RR389 pKa = 11.84RR390 pKa = 11.84ACVPFAPGTYY400 pKa = 10.06VILGDD405 pKa = 4.33DD406 pKa = 3.66VVIANDD412 pKa = 3.75KK413 pKa = 10.43VAEE416 pKa = 4.16QYY418 pKa = 8.64STLIRR423 pKa = 11.84WLGVEE428 pKa = 3.41ISMNKK433 pKa = 6.13THH435 pKa = 7.2RR436 pKa = 11.84SSHH439 pKa = 5.05SFEE442 pKa = 3.73IGKK445 pKa = 9.53RR446 pKa = 11.84WIHH449 pKa = 6.41RR450 pKa = 11.84GQEE453 pKa = 3.6ISPFSIVPWMQAKK466 pKa = 9.37NWAPAITQCIYY477 pKa = 10.66EE478 pKa = 4.19EE479 pKa = 4.63KK480 pKa = 10.04KK481 pKa = 10.81RR482 pKa = 11.84SWSLPFHH489 pKa = 6.12PVEE492 pKa = 4.38GVARR496 pKa = 11.84CILLQYY502 pKa = 9.86HH503 pKa = 6.86GKK505 pKa = 10.31LSSRR509 pKa = 11.84YY510 pKa = 8.54IDD512 pKa = 3.6HH513 pKa = 7.28LFDD516 pKa = 5.22LSQLNVAVYY525 pKa = 9.84HH526 pKa = 6.35SIKK529 pKa = 10.83EE530 pKa = 4.03MITWSDD536 pKa = 4.16CINIIAEE543 pKa = 4.24VFYY546 pKa = 9.4KK547 pKa = 9.79TKK549 pKa = 10.0WEE551 pKa = 4.12HH552 pKa = 6.75LDD554 pKa = 3.49RR555 pKa = 11.84LHH557 pKa = 6.92GRR559 pKa = 11.84LVQTYY564 pKa = 9.57VYY566 pKa = 10.86LRR568 pKa = 11.84LFNKK572 pKa = 9.88NAIPHH577 pKa = 5.98WSAIKK582 pKa = 10.36KK583 pKa = 10.14VILVEE588 pKa = 4.34NGFSMASIIQIIMGGRR604 pKa = 11.84SLSGASFSYY613 pKa = 11.39ADD615 pKa = 4.58LPHH618 pKa = 6.57KK619 pKa = 10.54KK620 pKa = 9.7IIRR623 pKa = 11.84RR624 pKa = 11.84YY625 pKa = 8.3AQEE628 pKa = 4.55FFDD631 pKa = 4.31KK632 pKa = 11.12NADD635 pKa = 3.61VVQGHH640 pKa = 5.4YY641 pKa = 10.68FEE643 pKa = 7.28DD644 pKa = 3.84GMLQRR649 pKa = 11.84EE650 pKa = 4.6AVRR653 pKa = 11.84SLGIPNVEE661 pKa = 4.02HH662 pKa = 7.27AFTKK666 pKa = 10.17SKK668 pKa = 10.22VRR670 pKa = 11.84RR671 pKa = 11.84EE672 pKa = 4.21VILSNNILKK681 pKa = 9.37EE682 pKa = 3.7LCSFVKK688 pKa = 10.36SVPSSGFTHH697 pKa = 6.99TSGSPVPTEE706 pKa = 3.26INEE709 pKa = 4.34YY710 pKa = 8.9IRR712 pKa = 11.84KK713 pKa = 8.31VNYY716 pKa = 9.8YY717 pKa = 10.05EE718 pKa = 4.01WDD720 pKa = 3.3TT721 pKa = 3.76
MM1 pKa = 7.44KK2 pKa = 10.63LIINIMKK9 pKa = 9.69IWAQSKK15 pKa = 10.17RR16 pKa = 11.84FLQIAKK22 pKa = 8.67TFYY25 pKa = 8.4TWYY28 pKa = 9.68SVYY31 pKa = 10.77YY32 pKa = 6.13PTCSPDD38 pKa = 4.25DD39 pKa = 3.21IAPFLEE45 pKa = 4.5RR46 pKa = 11.84VHH48 pKa = 6.88HH49 pKa = 6.03SLANRR54 pKa = 11.84GALATLEE61 pKa = 4.13WLKK64 pKa = 9.19EE65 pKa = 3.82TRR67 pKa = 11.84LAFTRR72 pKa = 11.84WICGDD77 pKa = 3.67SRR79 pKa = 11.84PDD81 pKa = 3.17VCVKK85 pKa = 10.42LDD87 pKa = 3.68SSGLPVMVKK96 pKa = 8.84STLGKK101 pKa = 10.5FIDD104 pKa = 3.93TARR107 pKa = 11.84GRR109 pKa = 11.84ALILTLLNCIRR120 pKa = 11.84SYY122 pKa = 11.33EE123 pKa = 4.09GLKK126 pKa = 10.37EE127 pKa = 3.93PSISDD132 pKa = 2.85ITSPYY137 pKa = 10.03KK138 pKa = 9.87GEE140 pKa = 3.84KK141 pKa = 9.38WIDD144 pKa = 3.52SYY146 pKa = 11.9LGLYY150 pKa = 8.75TKK152 pKa = 10.81DD153 pKa = 3.64FLRR156 pKa = 11.84KK157 pKa = 9.59YY158 pKa = 10.92GDD160 pKa = 2.92IHH162 pKa = 7.43RR163 pKa = 11.84PRR165 pKa = 11.84PQFKK169 pKa = 9.21QWLYY173 pKa = 4.77TTKK176 pKa = 10.37RR177 pKa = 11.84GPNGPSIATAYY188 pKa = 10.65HH189 pKa = 7.57DD190 pKa = 3.29ITLLTEE196 pKa = 3.45EE197 pKa = 5.03HH198 pKa = 5.69YY199 pKa = 10.7RR200 pKa = 11.84IYY202 pKa = 10.83EE203 pKa = 3.98VLAGKK208 pKa = 9.8DD209 pKa = 3.72YY210 pKa = 11.16KK211 pKa = 11.18DD212 pKa = 3.42SLLKK216 pKa = 10.4IKK218 pKa = 10.26EE219 pKa = 4.18IASDD223 pKa = 3.5PQEE226 pKa = 3.95FEE228 pKa = 4.9RR229 pKa = 11.84IRR231 pKa = 11.84KK232 pKa = 9.23DD233 pKa = 3.08FSLIVDD239 pKa = 3.8IKK241 pKa = 10.57SRR243 pKa = 11.84YY244 pKa = 6.84VEE246 pKa = 4.58KK247 pKa = 10.63ISVIPSPEE255 pKa = 3.51GKK257 pKa = 8.79MRR259 pKa = 11.84TISIASYY266 pKa = 8.65WAQNALDD273 pKa = 4.33PLHH276 pKa = 6.91RR277 pKa = 11.84CIFKK281 pKa = 10.37YY282 pKa = 10.27LRR284 pKa = 11.84KK285 pKa = 9.54IPNDD289 pKa = 2.89ITFDD293 pKa = 3.52QSKK296 pKa = 10.32GLSILNKK303 pKa = 9.66GQNDD307 pKa = 4.53SYY309 pKa = 11.11WSYY312 pKa = 11.6DD313 pKa = 3.23LKK315 pKa = 11.16AATDD319 pKa = 3.88RR320 pKa = 11.84FPRR323 pKa = 11.84STQTLLLSHH332 pKa = 7.51LIGQEE337 pKa = 4.35FANTWSLLISAEE349 pKa = 3.84HH350 pKa = 7.51DD351 pKa = 3.11IPAIGIRR358 pKa = 11.84AAWAVGQPIGKK369 pKa = 9.2LSSWAVFAYY378 pKa = 7.38THH380 pKa = 6.33HH381 pKa = 6.61FIIYY385 pKa = 7.55VAHH388 pKa = 6.99RR389 pKa = 11.84RR390 pKa = 11.84ACVPFAPGTYY400 pKa = 10.06VILGDD405 pKa = 4.33DD406 pKa = 3.66VVIANDD412 pKa = 3.75KK413 pKa = 10.43VAEE416 pKa = 4.16QYY418 pKa = 8.64STLIRR423 pKa = 11.84WLGVEE428 pKa = 3.41ISMNKK433 pKa = 6.13THH435 pKa = 7.2RR436 pKa = 11.84SSHH439 pKa = 5.05SFEE442 pKa = 3.73IGKK445 pKa = 9.53RR446 pKa = 11.84WIHH449 pKa = 6.41RR450 pKa = 11.84GQEE453 pKa = 3.6ISPFSIVPWMQAKK466 pKa = 9.37NWAPAITQCIYY477 pKa = 10.66EE478 pKa = 4.19EE479 pKa = 4.63KK480 pKa = 10.04KK481 pKa = 10.81RR482 pKa = 11.84SWSLPFHH489 pKa = 6.12PVEE492 pKa = 4.38GVARR496 pKa = 11.84CILLQYY502 pKa = 9.86HH503 pKa = 6.86GKK505 pKa = 10.31LSSRR509 pKa = 11.84YY510 pKa = 8.54IDD512 pKa = 3.6HH513 pKa = 7.28LFDD516 pKa = 5.22LSQLNVAVYY525 pKa = 9.84HH526 pKa = 6.35SIKK529 pKa = 10.83EE530 pKa = 4.03MITWSDD536 pKa = 4.16CINIIAEE543 pKa = 4.24VFYY546 pKa = 9.4KK547 pKa = 9.79TKK549 pKa = 10.0WEE551 pKa = 4.12HH552 pKa = 6.75LDD554 pKa = 3.49RR555 pKa = 11.84LHH557 pKa = 6.92GRR559 pKa = 11.84LVQTYY564 pKa = 9.57VYY566 pKa = 10.86LRR568 pKa = 11.84LFNKK572 pKa = 9.88NAIPHH577 pKa = 5.98WSAIKK582 pKa = 10.36KK583 pKa = 10.14VILVEE588 pKa = 4.34NGFSMASIIQIIMGGRR604 pKa = 11.84SLSGASFSYY613 pKa = 11.39ADD615 pKa = 4.58LPHH618 pKa = 6.57KK619 pKa = 10.54KK620 pKa = 9.7IIRR623 pKa = 11.84RR624 pKa = 11.84YY625 pKa = 8.3AQEE628 pKa = 4.55FFDD631 pKa = 4.31KK632 pKa = 11.12NADD635 pKa = 3.61VVQGHH640 pKa = 5.4YY641 pKa = 10.68FEE643 pKa = 7.28DD644 pKa = 3.84GMLQRR649 pKa = 11.84EE650 pKa = 4.6AVRR653 pKa = 11.84SLGIPNVEE661 pKa = 4.02HH662 pKa = 7.27AFTKK666 pKa = 10.17SKK668 pKa = 10.22VRR670 pKa = 11.84RR671 pKa = 11.84EE672 pKa = 4.21VILSNNILKK681 pKa = 9.37EE682 pKa = 3.7LCSFVKK688 pKa = 10.36SVPSSGFTHH697 pKa = 6.99TSGSPVPTEE706 pKa = 3.26INEE709 pKa = 4.34YY710 pKa = 8.9IRR712 pKa = 11.84KK713 pKa = 8.31VNYY716 pKa = 9.8YY717 pKa = 10.05EE718 pKa = 4.01WDD720 pKa = 3.3TT721 pKa = 3.76
Molecular weight: 83.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
721 |
721 |
721 |
721.0 |
83.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.38 ± 0.0 | 1.387 ± 0.0 |
4.854 ± 0.0 | 4.993 ± 0.0 |
4.161 ± 0.0 | 4.993 ± 0.0 |
3.467 ± 0.0 | 9.57 ± 0.0 |
6.935 ± 0.0 | 8.599 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.387 ± 0.0 | 3.19 ± 0.0 |
4.438 ± 0.0 | 3.051 ± 0.0 |
5.964 ± 0.0 | 8.599 ± 0.0 |
4.854 ± 0.0 | 5.409 ± 0.0 |
2.774 ± 0.0 | 4.993 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
