
Porcine coronavirus HKU15
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Deltacoronavirus; Buldecovirus; Coronavirus HKU15
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
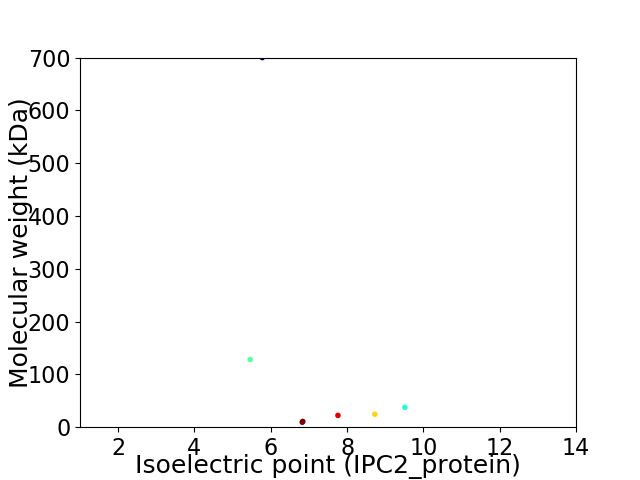
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
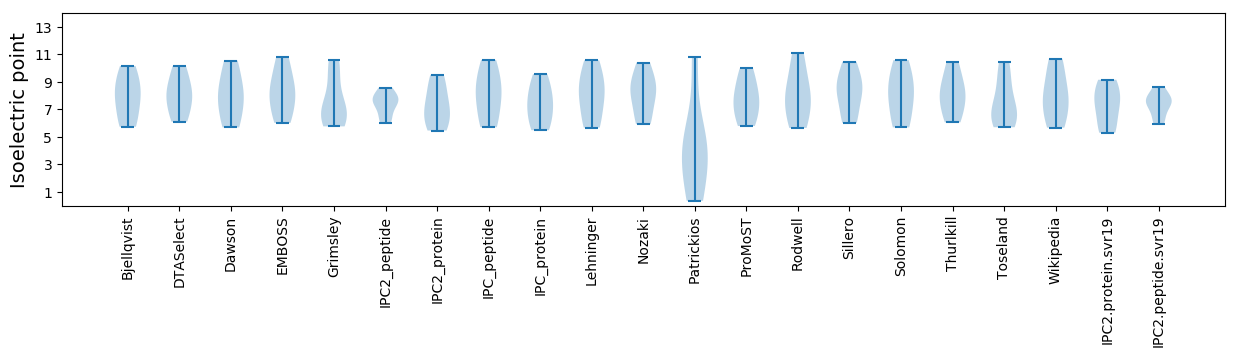
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9BQX9|H9BQX9_9NIDO Envelope protein OS=Porcine coronavirus HKU15 OX=1159905 GN=E PE=4 SV=1
MM1 pKa = 7.06QRR3 pKa = 11.84ALLIMTLLCLVRR15 pKa = 11.84AKK17 pKa = 10.61FADD20 pKa = 4.01DD21 pKa = 5.1LLDD24 pKa = 4.37LLTFPGAHH32 pKa = 6.45RR33 pKa = 11.84FLHH36 pKa = 5.95KK37 pKa = 9.15PTRR40 pKa = 11.84NSSSLYY46 pKa = 10.58SRR48 pKa = 11.84ANNNFDD54 pKa = 3.65VGVLPGYY61 pKa = 6.59PTKK64 pKa = 10.74NVNLFSPLTNSTLPINGLHH83 pKa = 7.3RR84 pKa = 11.84SYY86 pKa = 11.15QPLMLNCLTKK96 pKa = 9.63ITNHH100 pKa = 5.33TLSMYY105 pKa = 10.28LLPSEE110 pKa = 4.26IQTYY114 pKa = 9.47SCGGAMVKK122 pKa = 10.35YY123 pKa = 7.78QTHH126 pKa = 6.23DD127 pKa = 3.05AVRR130 pKa = 11.84IILDD134 pKa = 3.88LTATDD139 pKa = 4.67HH140 pKa = 7.17ISVEE144 pKa = 4.49VVGQHH149 pKa = 5.4GEE151 pKa = 3.98NYY153 pKa = 10.14VFVCSEE159 pKa = 3.71QFNYY163 pKa = 7.42TTALHH168 pKa = 6.36NSTFFSLNSEE178 pKa = 4.64LYY180 pKa = 10.4CFTNNTYY187 pKa = 10.84LGILPPDD194 pKa = 3.42LTDD197 pKa = 3.5FTVYY201 pKa = 9.24RR202 pKa = 11.84TGQFYY207 pKa = 11.3ANGYY211 pKa = 9.58LLGTLPITVNYY222 pKa = 8.41VRR224 pKa = 11.84LYY226 pKa = 10.42RR227 pKa = 11.84GHH229 pKa = 7.33LSANSAHH236 pKa = 6.5FALANLTDD244 pKa = 3.79TLITLTNTTISQITYY259 pKa = 9.11CDD261 pKa = 3.41KK262 pKa = 11.34SVVDD266 pKa = 5.56SIACQRR272 pKa = 11.84SSHH275 pKa = 5.05EE276 pKa = 4.23VEE278 pKa = 4.65DD279 pKa = 4.8GFYY282 pKa = 10.54SDD284 pKa = 3.98PKK286 pKa = 8.74SAVRR290 pKa = 11.84ARR292 pKa = 11.84QRR294 pKa = 11.84TIVTLPKK301 pKa = 10.35LPEE304 pKa = 4.28LEE306 pKa = 4.16VVQLNISAHH315 pKa = 5.23MDD317 pKa = 3.32FGEE320 pKa = 4.12ARR322 pKa = 11.84LDD324 pKa = 3.63SVTINGNTSYY334 pKa = 10.9CVTKK338 pKa = 10.17PYY340 pKa = 10.45FRR342 pKa = 11.84LEE344 pKa = 4.22TNFMCTGCTMNLRR357 pKa = 11.84TDD359 pKa = 3.25TCSFDD364 pKa = 3.98LSAVNNGMSFSQFCLSTEE382 pKa = 4.12SGACEE387 pKa = 3.57MKK389 pKa = 10.55IIVTYY394 pKa = 9.15VWNYY398 pKa = 10.41LLRR401 pKa = 11.84QRR403 pKa = 11.84LYY405 pKa = 8.2VTAVEE410 pKa = 4.68GQTHH414 pKa = 6.59TGTTSVHH421 pKa = 5.57ATDD424 pKa = 3.7TSSVITDD431 pKa = 3.13VCTDD435 pKa = 3.17YY436 pKa = 11.11TIYY439 pKa = 10.6GVSGTGIIKK448 pKa = 10.47PSDD451 pKa = 3.71LLLHH455 pKa = 6.34NGIAFTSPTGEE466 pKa = 3.83LYY468 pKa = 10.78AFKK471 pKa = 10.78NITTGKK477 pKa = 7.25TLQVLPCEE485 pKa = 4.59TPSQLIVINNTVVGAITSSNSTEE508 pKa = 3.87NNRR511 pKa = 11.84FTTTIVTPTFFYY523 pKa = 10.02STNDD527 pKa = 3.13TTFNCTKK534 pKa = 9.9PVLSYY539 pKa = 11.4GPISVCSDD547 pKa = 3.07GAIVGTSTLQNTRR560 pKa = 11.84PSIVSLYY567 pKa = 10.25DD568 pKa = 3.3GEE570 pKa = 4.75VEE572 pKa = 4.11IPSAFSLSVQTEE584 pKa = 4.19YY585 pKa = 11.31LQVQAEE591 pKa = 4.22QVIVDD596 pKa = 4.14CPQYY600 pKa = 10.72VCNGNSRR607 pKa = 11.84CLQLLAQYY615 pKa = 9.93TSACSNIEE623 pKa = 3.97AALHH627 pKa = 6.46SSAQLDD633 pKa = 3.48SRR635 pKa = 11.84EE636 pKa = 4.23IINMFKK642 pKa = 10.39TSTQSLQLANITNFKK657 pKa = 10.44GDD659 pKa = 3.79YY660 pKa = 9.12NFSSILTTRR669 pKa = 11.84IGGRR673 pKa = 11.84SAIEE677 pKa = 3.73DD678 pKa = 3.67LLFNKK683 pKa = 9.43VVTSGLGTVDD693 pKa = 3.9QDD695 pKa = 3.86YY696 pKa = 10.73KK697 pKa = 10.88SCSRR701 pKa = 11.84DD702 pKa = 3.16MAIADD707 pKa = 4.39LVCSQYY713 pKa = 11.52YY714 pKa = 9.28NGIMVLPGVVDD725 pKa = 3.77AEE727 pKa = 4.19KK728 pKa = 9.5MAMYY732 pKa = 9.35TGSLTGAMVFGGLTAAAAIPFATAVQARR760 pKa = 11.84LNYY763 pKa = 9.74VALQTNVLQEE773 pKa = 4.03NQKK776 pKa = 9.87ILAEE780 pKa = 4.27SFNQAVGNISLALSSVNDD798 pKa = 5.05AIQQTSEE805 pKa = 4.02ALNTVAIAIKK815 pKa = 10.22KK816 pKa = 8.47IQTVVNQQGEE826 pKa = 4.49ALSHH830 pKa = 5.94LTAQLSNNFQAISTSIQDD848 pKa = 2.88IYY850 pKa = 11.71NRR852 pKa = 11.84LEE854 pKa = 3.88EE855 pKa = 4.34VEE857 pKa = 5.42ANQQVDD863 pKa = 3.52RR864 pKa = 11.84LITGRR869 pKa = 11.84LAALNAYY876 pKa = 7.48VTQLLNQMSQIRR888 pKa = 11.84QSRR891 pKa = 11.84LLAQQKK897 pKa = 10.11INEE900 pKa = 4.5CVKK903 pKa = 10.38SQSSRR908 pKa = 11.84YY909 pKa = 8.35GFCGNGTHH917 pKa = 6.91IFSLTQTAPNGIFFMHH933 pKa = 7.03AVLVPNKK940 pKa = 7.88FTRR943 pKa = 11.84VNASAGICVDD953 pKa = 3.12NTRR956 pKa = 11.84GYY958 pKa = 10.79SLQPQLILYY967 pKa = 7.6QFNNSWRR974 pKa = 11.84VTPRR978 pKa = 11.84NMYY981 pKa = 9.92EE982 pKa = 3.71PRR984 pKa = 11.84LPRR987 pKa = 11.84QADD990 pKa = 4.83FIQLTDD996 pKa = 3.91CSVTFYY1002 pKa = 10.62NTTAANLPNIIPDD1015 pKa = 4.05IIDD1018 pKa = 3.39VNQTVSDD1025 pKa = 4.51IIDD1028 pKa = 4.04NLPTATPPQWDD1039 pKa = 3.32VGIYY1043 pKa = 10.91NNTILNLTVEE1053 pKa = 4.37INDD1056 pKa = 3.43LQEE1059 pKa = 3.91RR1060 pKa = 11.84SKK1062 pKa = 11.31NLSQIADD1069 pKa = 3.93RR1070 pKa = 11.84LQNYY1074 pKa = 9.14IDD1076 pKa = 3.98NLNNTLVDD1084 pKa = 4.3LEE1086 pKa = 4.17WLNRR1090 pKa = 11.84VEE1092 pKa = 5.99TYY1094 pKa = 10.72LKK1096 pKa = 8.97WPWYY1100 pKa = 8.19IWLAIALALIAFVTILITIFLCTGCCGGCFGCCGGCFGLFSKK1142 pKa = 10.53KK1143 pKa = 9.65KK1144 pKa = 10.16RR1145 pKa = 11.84YY1146 pKa = 8.31TDD1148 pKa = 3.81DD1149 pKa = 3.27QPTPSFKK1156 pKa = 10.66FKK1158 pKa = 10.4EE1159 pKa = 4.19WW1160 pKa = 3.21
MM1 pKa = 7.06QRR3 pKa = 11.84ALLIMTLLCLVRR15 pKa = 11.84AKK17 pKa = 10.61FADD20 pKa = 4.01DD21 pKa = 5.1LLDD24 pKa = 4.37LLTFPGAHH32 pKa = 6.45RR33 pKa = 11.84FLHH36 pKa = 5.95KK37 pKa = 9.15PTRR40 pKa = 11.84NSSSLYY46 pKa = 10.58SRR48 pKa = 11.84ANNNFDD54 pKa = 3.65VGVLPGYY61 pKa = 6.59PTKK64 pKa = 10.74NVNLFSPLTNSTLPINGLHH83 pKa = 7.3RR84 pKa = 11.84SYY86 pKa = 11.15QPLMLNCLTKK96 pKa = 9.63ITNHH100 pKa = 5.33TLSMYY105 pKa = 10.28LLPSEE110 pKa = 4.26IQTYY114 pKa = 9.47SCGGAMVKK122 pKa = 10.35YY123 pKa = 7.78QTHH126 pKa = 6.23DD127 pKa = 3.05AVRR130 pKa = 11.84IILDD134 pKa = 3.88LTATDD139 pKa = 4.67HH140 pKa = 7.17ISVEE144 pKa = 4.49VVGQHH149 pKa = 5.4GEE151 pKa = 3.98NYY153 pKa = 10.14VFVCSEE159 pKa = 3.71QFNYY163 pKa = 7.42TTALHH168 pKa = 6.36NSTFFSLNSEE178 pKa = 4.64LYY180 pKa = 10.4CFTNNTYY187 pKa = 10.84LGILPPDD194 pKa = 3.42LTDD197 pKa = 3.5FTVYY201 pKa = 9.24RR202 pKa = 11.84TGQFYY207 pKa = 11.3ANGYY211 pKa = 9.58LLGTLPITVNYY222 pKa = 8.41VRR224 pKa = 11.84LYY226 pKa = 10.42RR227 pKa = 11.84GHH229 pKa = 7.33LSANSAHH236 pKa = 6.5FALANLTDD244 pKa = 3.79TLITLTNTTISQITYY259 pKa = 9.11CDD261 pKa = 3.41KK262 pKa = 11.34SVVDD266 pKa = 5.56SIACQRR272 pKa = 11.84SSHH275 pKa = 5.05EE276 pKa = 4.23VEE278 pKa = 4.65DD279 pKa = 4.8GFYY282 pKa = 10.54SDD284 pKa = 3.98PKK286 pKa = 8.74SAVRR290 pKa = 11.84ARR292 pKa = 11.84QRR294 pKa = 11.84TIVTLPKK301 pKa = 10.35LPEE304 pKa = 4.28LEE306 pKa = 4.16VVQLNISAHH315 pKa = 5.23MDD317 pKa = 3.32FGEE320 pKa = 4.12ARR322 pKa = 11.84LDD324 pKa = 3.63SVTINGNTSYY334 pKa = 10.9CVTKK338 pKa = 10.17PYY340 pKa = 10.45FRR342 pKa = 11.84LEE344 pKa = 4.22TNFMCTGCTMNLRR357 pKa = 11.84TDD359 pKa = 3.25TCSFDD364 pKa = 3.98LSAVNNGMSFSQFCLSTEE382 pKa = 4.12SGACEE387 pKa = 3.57MKK389 pKa = 10.55IIVTYY394 pKa = 9.15VWNYY398 pKa = 10.41LLRR401 pKa = 11.84QRR403 pKa = 11.84LYY405 pKa = 8.2VTAVEE410 pKa = 4.68GQTHH414 pKa = 6.59TGTTSVHH421 pKa = 5.57ATDD424 pKa = 3.7TSSVITDD431 pKa = 3.13VCTDD435 pKa = 3.17YY436 pKa = 11.11TIYY439 pKa = 10.6GVSGTGIIKK448 pKa = 10.47PSDD451 pKa = 3.71LLLHH455 pKa = 6.34NGIAFTSPTGEE466 pKa = 3.83LYY468 pKa = 10.78AFKK471 pKa = 10.78NITTGKK477 pKa = 7.25TLQVLPCEE485 pKa = 4.59TPSQLIVINNTVVGAITSSNSTEE508 pKa = 3.87NNRR511 pKa = 11.84FTTTIVTPTFFYY523 pKa = 10.02STNDD527 pKa = 3.13TTFNCTKK534 pKa = 9.9PVLSYY539 pKa = 11.4GPISVCSDD547 pKa = 3.07GAIVGTSTLQNTRR560 pKa = 11.84PSIVSLYY567 pKa = 10.25DD568 pKa = 3.3GEE570 pKa = 4.75VEE572 pKa = 4.11IPSAFSLSVQTEE584 pKa = 4.19YY585 pKa = 11.31LQVQAEE591 pKa = 4.22QVIVDD596 pKa = 4.14CPQYY600 pKa = 10.72VCNGNSRR607 pKa = 11.84CLQLLAQYY615 pKa = 9.93TSACSNIEE623 pKa = 3.97AALHH627 pKa = 6.46SSAQLDD633 pKa = 3.48SRR635 pKa = 11.84EE636 pKa = 4.23IINMFKK642 pKa = 10.39TSTQSLQLANITNFKK657 pKa = 10.44GDD659 pKa = 3.79YY660 pKa = 9.12NFSSILTTRR669 pKa = 11.84IGGRR673 pKa = 11.84SAIEE677 pKa = 3.73DD678 pKa = 3.67LLFNKK683 pKa = 9.43VVTSGLGTVDD693 pKa = 3.9QDD695 pKa = 3.86YY696 pKa = 10.73KK697 pKa = 10.88SCSRR701 pKa = 11.84DD702 pKa = 3.16MAIADD707 pKa = 4.39LVCSQYY713 pKa = 11.52YY714 pKa = 9.28NGIMVLPGVVDD725 pKa = 3.77AEE727 pKa = 4.19KK728 pKa = 9.5MAMYY732 pKa = 9.35TGSLTGAMVFGGLTAAAAIPFATAVQARR760 pKa = 11.84LNYY763 pKa = 9.74VALQTNVLQEE773 pKa = 4.03NQKK776 pKa = 9.87ILAEE780 pKa = 4.27SFNQAVGNISLALSSVNDD798 pKa = 5.05AIQQTSEE805 pKa = 4.02ALNTVAIAIKK815 pKa = 10.22KK816 pKa = 8.47IQTVVNQQGEE826 pKa = 4.49ALSHH830 pKa = 5.94LTAQLSNNFQAISTSIQDD848 pKa = 2.88IYY850 pKa = 11.71NRR852 pKa = 11.84LEE854 pKa = 3.88EE855 pKa = 4.34VEE857 pKa = 5.42ANQQVDD863 pKa = 3.52RR864 pKa = 11.84LITGRR869 pKa = 11.84LAALNAYY876 pKa = 7.48VTQLLNQMSQIRR888 pKa = 11.84QSRR891 pKa = 11.84LLAQQKK897 pKa = 10.11INEE900 pKa = 4.5CVKK903 pKa = 10.38SQSSRR908 pKa = 11.84YY909 pKa = 8.35GFCGNGTHH917 pKa = 6.91IFSLTQTAPNGIFFMHH933 pKa = 7.03AVLVPNKK940 pKa = 7.88FTRR943 pKa = 11.84VNASAGICVDD953 pKa = 3.12NTRR956 pKa = 11.84GYY958 pKa = 10.79SLQPQLILYY967 pKa = 7.6QFNNSWRR974 pKa = 11.84VTPRR978 pKa = 11.84NMYY981 pKa = 9.92EE982 pKa = 3.71PRR984 pKa = 11.84LPRR987 pKa = 11.84QADD990 pKa = 4.83FIQLTDD996 pKa = 3.91CSVTFYY1002 pKa = 10.62NTTAANLPNIIPDD1015 pKa = 4.05IIDD1018 pKa = 3.39VNQTVSDD1025 pKa = 4.51IIDD1028 pKa = 4.04NLPTATPPQWDD1039 pKa = 3.32VGIYY1043 pKa = 10.91NNTILNLTVEE1053 pKa = 4.37INDD1056 pKa = 3.43LQEE1059 pKa = 3.91RR1060 pKa = 11.84SKK1062 pKa = 11.31NLSQIADD1069 pKa = 3.93RR1070 pKa = 11.84LQNYY1074 pKa = 9.14IDD1076 pKa = 3.98NLNNTLVDD1084 pKa = 4.3LEE1086 pKa = 4.17WLNRR1090 pKa = 11.84VEE1092 pKa = 5.99TYY1094 pKa = 10.72LKK1096 pKa = 8.97WPWYY1100 pKa = 8.19IWLAIALALIAFVTILITIFLCTGCCGGCFGCCGGCFGLFSKK1142 pKa = 10.53KK1143 pKa = 9.65KK1144 pKa = 10.16RR1145 pKa = 11.84YY1146 pKa = 8.31TDD1148 pKa = 3.81DD1149 pKa = 3.27QPTPSFKK1156 pKa = 10.66FKK1158 pKa = 10.4EE1159 pKa = 4.19WW1160 pKa = 3.21
Molecular weight: 128.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A060A825|A0A060A825_9NIDO Spike glycoprotein OS=Porcine coronavirus HKU15 OX=1159905 GN=S PE=4 SV=1
MM1 pKa = 7.64AAPVVPTTDD10 pKa = 3.28ASWFQVLKK18 pKa = 10.58AQNKK22 pKa = 9.03KK23 pKa = 8.6ATHH26 pKa = 5.19PQFRR30 pKa = 11.84GNGVPLNSAIKK41 pKa = 9.29PVEE44 pKa = 3.72NHH46 pKa = 6.56GYY48 pKa = 7.02WLRR51 pKa = 11.84YY52 pKa = 7.01TRR54 pKa = 11.84QKK56 pKa = 10.75PGGTPIPPSYY66 pKa = 10.88AFYY69 pKa = 9.95YY70 pKa = 9.86TGTGPRR76 pKa = 11.84GNLKK80 pKa = 10.6YY81 pKa = 11.01GEE83 pKa = 4.96LPPNDD88 pKa = 3.78TPATTRR94 pKa = 11.84VTWVKK99 pKa = 10.94GSGADD104 pKa = 3.53TSIKK108 pKa = 9.78PHH110 pKa = 4.45VAKK113 pKa = 10.68RR114 pKa = 11.84NPNNPKK120 pKa = 9.85HH121 pKa = 5.96QLLPLRR127 pKa = 11.84FPTGDD132 pKa = 3.2GPAQGFRR139 pKa = 11.84VDD141 pKa = 3.64PFNARR146 pKa = 11.84GRR148 pKa = 11.84PQEE151 pKa = 4.34RR152 pKa = 11.84GSGPRR157 pKa = 11.84SQSVNSRR164 pKa = 11.84GTGNQPRR171 pKa = 11.84KK172 pKa = 9.37RR173 pKa = 11.84DD174 pKa = 3.44QSAPAAVRR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 7.09TQHH187 pKa = 5.18QAPKK191 pKa = 9.09RR192 pKa = 11.84TLPKK196 pKa = 10.35GKK198 pKa = 8.69TISQVFGNRR207 pKa = 11.84SRR209 pKa = 11.84TGANVGSADD218 pKa = 3.79TEE220 pKa = 4.29KK221 pKa = 10.47TGMADD226 pKa = 3.45PRR228 pKa = 11.84IMALARR234 pKa = 11.84HH235 pKa = 5.28VPGVQEE241 pKa = 3.98MLFAGHH247 pKa = 7.25LEE249 pKa = 4.47SNFQAGAITLTFSYY263 pKa = 10.44SITVKK268 pKa = 10.44EE269 pKa = 4.26GSPDD273 pKa = 3.46YY274 pKa = 11.25DD275 pKa = 3.37RR276 pKa = 11.84LKK278 pKa = 10.99DD279 pKa = 3.64ALNTVVNQTYY289 pKa = 9.4EE290 pKa = 4.24PPTKK294 pKa = 8.96PTKK297 pKa = 10.21DD298 pKa = 3.46KK299 pKa = 11.42KK300 pKa = 10.17PDD302 pKa = 3.57KK303 pKa = 10.07QDD305 pKa = 3.05QSAKK309 pKa = 10.09PKK311 pKa = 8.42QQKK314 pKa = 9.59KK315 pKa = 8.18PKK317 pKa = 9.94KK318 pKa = 8.31VTLPADD324 pKa = 3.66KK325 pKa = 10.57QDD327 pKa = 3.86WEE329 pKa = 4.04WDD331 pKa = 3.38DD332 pKa = 3.93AFEE335 pKa = 4.83IKK337 pKa = 10.2QEE339 pKa = 4.19SAAA342 pKa = 4.0
MM1 pKa = 7.64AAPVVPTTDD10 pKa = 3.28ASWFQVLKK18 pKa = 10.58AQNKK22 pKa = 9.03KK23 pKa = 8.6ATHH26 pKa = 5.19PQFRR30 pKa = 11.84GNGVPLNSAIKK41 pKa = 9.29PVEE44 pKa = 3.72NHH46 pKa = 6.56GYY48 pKa = 7.02WLRR51 pKa = 11.84YY52 pKa = 7.01TRR54 pKa = 11.84QKK56 pKa = 10.75PGGTPIPPSYY66 pKa = 10.88AFYY69 pKa = 9.95YY70 pKa = 9.86TGTGPRR76 pKa = 11.84GNLKK80 pKa = 10.6YY81 pKa = 11.01GEE83 pKa = 4.96LPPNDD88 pKa = 3.78TPATTRR94 pKa = 11.84VTWVKK99 pKa = 10.94GSGADD104 pKa = 3.53TSIKK108 pKa = 9.78PHH110 pKa = 4.45VAKK113 pKa = 10.68RR114 pKa = 11.84NPNNPKK120 pKa = 9.85HH121 pKa = 5.96QLLPLRR127 pKa = 11.84FPTGDD132 pKa = 3.2GPAQGFRR139 pKa = 11.84VDD141 pKa = 3.64PFNARR146 pKa = 11.84GRR148 pKa = 11.84PQEE151 pKa = 4.34RR152 pKa = 11.84GSGPRR157 pKa = 11.84SQSVNSRR164 pKa = 11.84GTGNQPRR171 pKa = 11.84KK172 pKa = 9.37RR173 pKa = 11.84DD174 pKa = 3.44QSAPAAVRR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 7.09TQHH187 pKa = 5.18QAPKK191 pKa = 9.09RR192 pKa = 11.84TLPKK196 pKa = 10.35GKK198 pKa = 8.69TISQVFGNRR207 pKa = 11.84SRR209 pKa = 11.84TGANVGSADD218 pKa = 3.79TEE220 pKa = 4.29KK221 pKa = 10.47TGMADD226 pKa = 3.45PRR228 pKa = 11.84IMALARR234 pKa = 11.84HH235 pKa = 5.28VPGVQEE241 pKa = 3.98MLFAGHH247 pKa = 7.25LEE249 pKa = 4.47SNFQAGAITLTFSYY263 pKa = 10.44SITVKK268 pKa = 10.44EE269 pKa = 4.26GSPDD273 pKa = 3.46YY274 pKa = 11.25DD275 pKa = 3.37RR276 pKa = 11.84LKK278 pKa = 10.99DD279 pKa = 3.64ALNTVVNQTYY289 pKa = 9.4EE290 pKa = 4.24PPTKK294 pKa = 8.96PTKK297 pKa = 10.21DD298 pKa = 3.46KK299 pKa = 11.42KK300 pKa = 10.17PDD302 pKa = 3.57KK303 pKa = 10.07QDD305 pKa = 3.05QSAKK309 pKa = 10.09PKK311 pKa = 8.42QQKK314 pKa = 9.59KK315 pKa = 8.18PKK317 pKa = 9.94KK318 pKa = 8.31VTLPADD324 pKa = 3.66KK325 pKa = 10.57QDD327 pKa = 3.86WEE329 pKa = 4.04WDD331 pKa = 3.38DD332 pKa = 3.93AFEE335 pKa = 4.83IKK337 pKa = 10.2QEE339 pKa = 4.19SAAA342 pKa = 4.0
Molecular weight: 37.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8363 |
83 |
6267 |
1194.7 |
133.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.701 ± 0.453 | 2.487 ± 0.425 |
5.285 ± 0.556 | 4.054 ± 0.292 |
4.341 ± 0.429 | 4.938 ± 0.511 |
2.344 ± 0.305 | 5.991 ± 0.676 |
4.938 ± 0.761 | 9.59 ± 1.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.818 ± 0.316 | 5.536 ± 0.617 |
5.034 ± 1.232 | 4.4 ± 0.46 |
3.802 ± 0.703 | 6.11 ± 0.589 |
7.988 ± 0.869 | 8.095 ± 0.596 |
0.849 ± 0.273 | 4.699 ± 0.546 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
