
Giant panda polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus; Ailuropoda melanoleuca polyomavirus 1
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
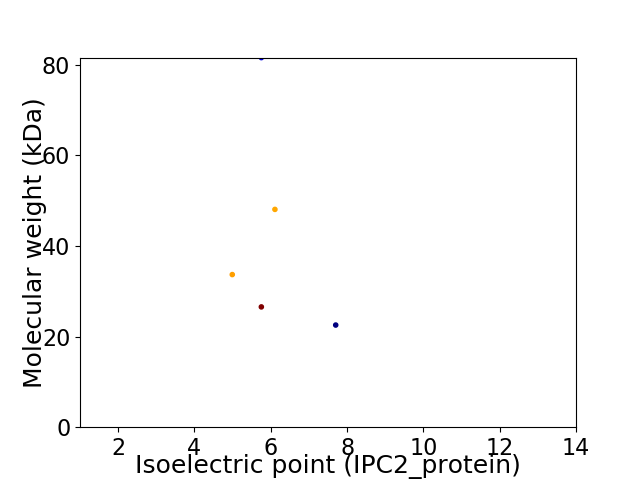
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

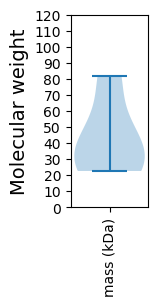
Protein with the lowest isoelectric point:
>tr|A0A220IG73|A0A220IG73_9POLY Minor capsid protein OS=Giant panda polyomavirus OX=2016463 PE=3 SV=1
MM1 pKa = 7.47GGVLSIVVDD10 pKa = 4.58LFTLLPEE17 pKa = 4.81LSASTGFGIEE27 pKa = 4.44AILAGEE33 pKa = 4.15AAASVEE39 pKa = 4.07AQVASLMLIEE49 pKa = 4.55TMGPLDD55 pKa = 4.02ALASLGLSAEE65 pKa = 4.38TFSLLSAMPGFFQDD79 pKa = 3.17IVGLGVLFQTVSGASSLVAAGIQLGRR105 pKa = 11.84HH106 pKa = 4.89EE107 pKa = 4.42VSVVNRR113 pKa = 11.84NMALVPWIPQDD124 pKa = 3.57LFDD127 pKa = 4.7IYY129 pKa = 11.02FPGVSSFSYY138 pKa = 10.26VVNIVADD145 pKa = 3.38WGEE148 pKa = 4.32SLFHH152 pKa = 6.61SLARR156 pKa = 11.84NIWDD160 pKa = 3.25ALMRR164 pKa = 11.84EE165 pKa = 4.09SRR167 pKa = 11.84AQITHH172 pKa = 6.17ATRR175 pKa = 11.84EE176 pKa = 4.05LTLRR180 pKa = 11.84GVQTFQDD187 pKa = 3.04TMARR191 pKa = 11.84VIEE194 pKa = 3.99RR195 pKa = 11.84ARR197 pKa = 11.84WVVTNGPVHH206 pKa = 7.02AYY208 pKa = 10.0SAIEE212 pKa = 4.08EE213 pKa = 4.67YY214 pKa = 10.93YY215 pKa = 10.53RR216 pKa = 11.84ALPPINPAQARR227 pKa = 11.84QLARR231 pKa = 11.84RR232 pKa = 11.84LEE234 pKa = 4.22IKK236 pKa = 9.68TPEE239 pKa = 4.1RR240 pKa = 11.84LTIEE244 pKa = 4.17SPEE247 pKa = 3.9EE248 pKa = 3.87SGEE251 pKa = 4.08VVQRR255 pKa = 11.84YY256 pKa = 7.11EE257 pKa = 3.98PPGGAFQRR265 pKa = 11.84ATPDD269 pKa = 2.53WMLPLILGLYY279 pKa = 10.53GDD281 pKa = 4.65ITPTWSSYY289 pKa = 10.81LEE291 pKa = 4.48HH292 pKa = 8.12IEE294 pKa = 4.84AEE296 pKa = 3.86EE297 pKa = 4.62DD298 pKa = 3.82GPKK301 pKa = 10.08KK302 pKa = 10.18KK303 pKa = 9.95RR304 pKa = 11.84RR305 pKa = 11.84KK306 pKa = 7.86QQ307 pKa = 3.16
MM1 pKa = 7.47GGVLSIVVDD10 pKa = 4.58LFTLLPEE17 pKa = 4.81LSASTGFGIEE27 pKa = 4.44AILAGEE33 pKa = 4.15AAASVEE39 pKa = 4.07AQVASLMLIEE49 pKa = 4.55TMGPLDD55 pKa = 4.02ALASLGLSAEE65 pKa = 4.38TFSLLSAMPGFFQDD79 pKa = 3.17IVGLGVLFQTVSGASSLVAAGIQLGRR105 pKa = 11.84HH106 pKa = 4.89EE107 pKa = 4.42VSVVNRR113 pKa = 11.84NMALVPWIPQDD124 pKa = 3.57LFDD127 pKa = 4.7IYY129 pKa = 11.02FPGVSSFSYY138 pKa = 10.26VVNIVADD145 pKa = 3.38WGEE148 pKa = 4.32SLFHH152 pKa = 6.61SLARR156 pKa = 11.84NIWDD160 pKa = 3.25ALMRR164 pKa = 11.84EE165 pKa = 4.09SRR167 pKa = 11.84AQITHH172 pKa = 6.17ATRR175 pKa = 11.84EE176 pKa = 4.05LTLRR180 pKa = 11.84GVQTFQDD187 pKa = 3.04TMARR191 pKa = 11.84VIEE194 pKa = 3.99RR195 pKa = 11.84ARR197 pKa = 11.84WVVTNGPVHH206 pKa = 7.02AYY208 pKa = 10.0SAIEE212 pKa = 4.08EE213 pKa = 4.67YY214 pKa = 10.93YY215 pKa = 10.53RR216 pKa = 11.84ALPPINPAQARR227 pKa = 11.84QLARR231 pKa = 11.84RR232 pKa = 11.84LEE234 pKa = 4.22IKK236 pKa = 9.68TPEE239 pKa = 4.1RR240 pKa = 11.84LTIEE244 pKa = 4.17SPEE247 pKa = 3.9EE248 pKa = 3.87SGEE251 pKa = 4.08VVQRR255 pKa = 11.84YY256 pKa = 7.11EE257 pKa = 3.98PPGGAFQRR265 pKa = 11.84ATPDD269 pKa = 2.53WMLPLILGLYY279 pKa = 10.53GDD281 pKa = 4.65ITPTWSSYY289 pKa = 10.81LEE291 pKa = 4.48HH292 pKa = 8.12IEE294 pKa = 4.84AEE296 pKa = 3.86EE297 pKa = 4.62DD298 pKa = 3.82GPKK301 pKa = 10.08KK302 pKa = 10.18KK303 pKa = 9.95RR304 pKa = 11.84RR305 pKa = 11.84KK306 pKa = 7.86QQ307 pKa = 3.16
Molecular weight: 33.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IG84|A0A220IG84_9POLY Small T antigen OS=Giant panda polyomavirus OX=2016463 PE=4 SV=1
MM1 pKa = 7.97DD2 pKa = 4.93AVLTKK7 pKa = 10.6AEE9 pKa = 4.3KK10 pKa = 10.57DD11 pKa = 3.44EE12 pKa = 4.29LMSLLGLAPTCYY24 pKa = 10.84GNLPLMQQKK33 pKa = 9.71YY34 pKa = 9.84KK35 pKa = 10.33KK36 pKa = 10.17ASLKK40 pKa = 9.45FHH42 pKa = 7.18PDD44 pKa = 2.82KK45 pKa = 11.61GGDD48 pKa = 3.55EE49 pKa = 4.35EE50 pKa = 4.93KK51 pKa = 10.28MKK53 pKa = 10.72RR54 pKa = 11.84LNCLFGKK61 pKa = 10.22VYY63 pKa = 11.07NSFSDD68 pKa = 5.19LRR70 pKa = 11.84DD71 pKa = 3.43QPRR74 pKa = 11.84GSCSSQVNLWEE85 pKa = 4.19KK86 pKa = 9.82TVITTADD93 pKa = 3.32FFGKK97 pKa = 10.15KK98 pKa = 8.62FEE100 pKa = 4.61KK101 pKa = 10.71NCVKK105 pKa = 10.31HH106 pKa = 4.75YY107 pKa = 10.16HH108 pKa = 5.05YY109 pKa = 10.62CILEE113 pKa = 4.16GLSKK117 pKa = 11.19YY118 pKa = 7.4CTCICCLLDD127 pKa = 3.74QQHH130 pKa = 6.24HH131 pKa = 4.27QLKK134 pKa = 9.9RR135 pKa = 11.84KK136 pKa = 9.28KK137 pKa = 10.39SKK139 pKa = 10.09PCLVWGEE146 pKa = 4.26CLCFNCYY153 pKa = 10.0RR154 pKa = 11.84SWFGLDD160 pKa = 3.93LNTEE164 pKa = 4.21TLHH167 pKa = 4.91WWKK170 pKa = 10.48FCIYY174 pKa = 9.17RR175 pKa = 11.84LPMEE179 pKa = 4.52WLNILGDD186 pKa = 3.75IKK188 pKa = 11.12LSDD191 pKa = 3.23WW192 pKa = 3.63
MM1 pKa = 7.97DD2 pKa = 4.93AVLTKK7 pKa = 10.6AEE9 pKa = 4.3KK10 pKa = 10.57DD11 pKa = 3.44EE12 pKa = 4.29LMSLLGLAPTCYY24 pKa = 10.84GNLPLMQQKK33 pKa = 9.71YY34 pKa = 9.84KK35 pKa = 10.33KK36 pKa = 10.17ASLKK40 pKa = 9.45FHH42 pKa = 7.18PDD44 pKa = 2.82KK45 pKa = 11.61GGDD48 pKa = 3.55EE49 pKa = 4.35EE50 pKa = 4.93KK51 pKa = 10.28MKK53 pKa = 10.72RR54 pKa = 11.84LNCLFGKK61 pKa = 10.22VYY63 pKa = 11.07NSFSDD68 pKa = 5.19LRR70 pKa = 11.84DD71 pKa = 3.43QPRR74 pKa = 11.84GSCSSQVNLWEE85 pKa = 4.19KK86 pKa = 9.82TVITTADD93 pKa = 3.32FFGKK97 pKa = 10.15KK98 pKa = 8.62FEE100 pKa = 4.61KK101 pKa = 10.71NCVKK105 pKa = 10.31HH106 pKa = 4.75YY107 pKa = 10.16HH108 pKa = 5.05YY109 pKa = 10.62CILEE113 pKa = 4.16GLSKK117 pKa = 11.19YY118 pKa = 7.4CTCICCLLDD127 pKa = 3.74QQHH130 pKa = 6.24HH131 pKa = 4.27QLKK134 pKa = 9.9RR135 pKa = 11.84KK136 pKa = 9.28KK137 pKa = 10.39SKK139 pKa = 10.09PCLVWGEE146 pKa = 4.26CLCFNCYY153 pKa = 10.0RR154 pKa = 11.84SWFGLDD160 pKa = 3.93LNTEE164 pKa = 4.21TLHH167 pKa = 4.91WWKK170 pKa = 10.48FCIYY174 pKa = 9.17RR175 pKa = 11.84LPMEE179 pKa = 4.52WLNILGDD186 pKa = 3.75IKK188 pKa = 11.12LSDD191 pKa = 3.23WW192 pKa = 3.63
Molecular weight: 22.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1899 |
192 |
723 |
379.8 |
42.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.161 ± 1.18 | 2.37 ± 0.865 |
5.477 ± 0.774 | 5.951 ± 0.591 |
3.739 ± 0.422 | 6.108 ± 0.597 |
1.685 ± 0.273 | 4.739 ± 0.516 |
6.161 ± 1.549 | 9.637 ± 0.892 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.844 ± 0.274 | 3.739 ± 0.525 |
7.846 ± 1.093 | 4.16 ± 0.185 |
4.476 ± 0.839 | 7.794 ± 0.463 |
5.74 ± 0.543 | 6.108 ± 0.753 |
1.58 ± 0.448 | 3.686 ± 0.347 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
