
Vibrio phage VP-HS15
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Colwellvirinae; Kaohsiungvirus; unclassified Kaohsiungvirus
Average proteome isoelectric point is 5.66
Get precalculated fractions of proteins
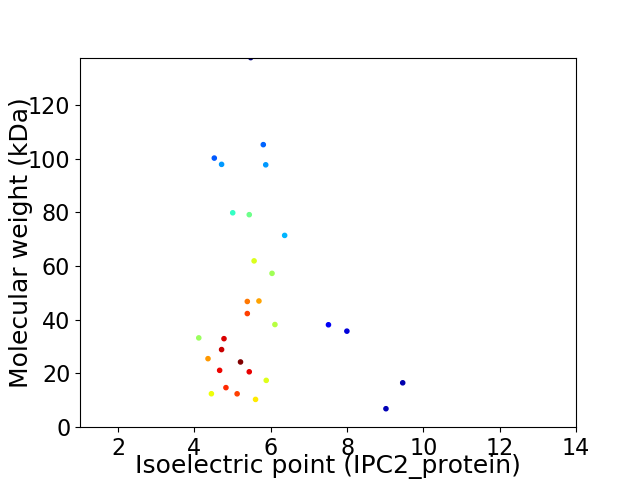
Virtual 2D-PAGE plot for 30 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9LT58|A0A6B9LT58_9CAUD Uncharacterized protein OS=Vibrio phage VP-HS15 OX=2686284 PE=4 SV=1
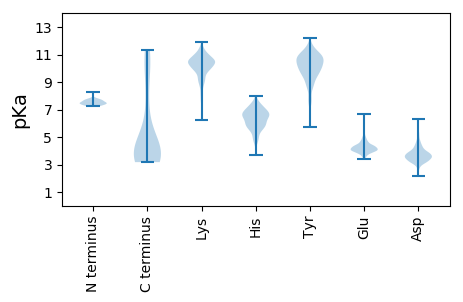
MM1 pKa = 7.73DD2 pKa = 4.72VEE4 pKa = 5.27IIDD7 pKa = 3.95YY8 pKa = 11.06NSTDD12 pKa = 3.54VVDD15 pKa = 3.92LTGGKK20 pKa = 7.73TEE22 pKa = 4.58PEE24 pKa = 4.09DD25 pKa = 3.48PKK27 pKa = 10.94QEE29 pKa = 4.02QEE31 pKa = 4.25PEE33 pKa = 3.94QSTVTEE39 pKa = 4.0PVEE42 pKa = 3.96PAEE45 pKa = 4.49EE46 pKa = 4.64SPASTPAEE54 pKa = 4.22DD55 pKa = 5.0ADD57 pKa = 3.97AEE59 pKa = 4.48SAATEE64 pKa = 4.12PEE66 pKa = 4.02KK67 pKa = 10.57TEE69 pKa = 3.75QEE71 pKa = 4.15EE72 pKa = 4.46PEE74 pKa = 4.16EE75 pKa = 4.41SGEE78 pKa = 4.0KK79 pKa = 10.0PEE81 pKa = 4.19YY82 pKa = 10.21FFNGEE87 pKa = 4.03AVSVEE92 pKa = 4.04VPEE95 pKa = 4.28EE96 pKa = 3.63VRR98 pKa = 11.84NALKK102 pKa = 10.28EE103 pKa = 3.98AEE105 pKa = 4.12IDD107 pKa = 3.74EE108 pKa = 4.62KK109 pKa = 11.59ALLSQLFKK117 pKa = 10.97KK118 pKa = 10.75DD119 pKa = 3.18GDD121 pKa = 3.77FSLDD125 pKa = 3.12EE126 pKa = 4.0EE127 pKa = 4.82TRR129 pKa = 11.84TKK131 pKa = 10.96LEE133 pKa = 4.21SKK135 pKa = 10.39FGKK138 pKa = 8.56TLVDD142 pKa = 4.87GYY144 pKa = 11.49LNMYY148 pKa = 10.45KK149 pKa = 10.57GMNEE153 pKa = 3.91QAIKK157 pKa = 10.39AAAADD162 pKa = 3.97KK163 pKa = 9.71EE164 pKa = 4.37QQEE167 pKa = 4.71VTSKK171 pKa = 10.1QQAEE175 pKa = 4.21EE176 pKa = 4.1FMTTVGGEE184 pKa = 4.03EE185 pKa = 4.36GLVAMEE191 pKa = 5.82DD192 pKa = 4.36FILKK196 pKa = 10.28NFDD199 pKa = 3.74DD200 pKa = 4.43KK201 pKa = 11.56QIAAYY206 pKa = 8.87NAVMEE211 pKa = 4.7SEE213 pKa = 4.2SHH215 pKa = 5.2EE216 pKa = 4.04QQLLIIQTVKK226 pKa = 11.12AQMEE230 pKa = 4.37LSDD233 pKa = 4.3KK234 pKa = 10.6LQNGDD239 pKa = 4.13KK240 pKa = 10.54NIKK243 pKa = 10.25LLGDD247 pKa = 4.04DD248 pKa = 4.16TSSTPTGTPLDD259 pKa = 3.86KK260 pKa = 11.11GYY262 pKa = 8.86LTSDD266 pKa = 3.7EE267 pKa = 4.59FQAIMDD273 pKa = 3.96SDD275 pKa = 4.22KK276 pKa = 10.55YY277 pKa = 8.96WTDD280 pKa = 3.51FEE282 pKa = 4.7YY283 pKa = 9.48QTKK286 pKa = 10.19VDD288 pKa = 3.71AARR291 pKa = 11.84SAGLKK296 pKa = 10.43RR297 pKa = 11.84NLL299 pKa = 3.73
MM1 pKa = 7.73DD2 pKa = 4.72VEE4 pKa = 5.27IIDD7 pKa = 3.95YY8 pKa = 11.06NSTDD12 pKa = 3.54VVDD15 pKa = 3.92LTGGKK20 pKa = 7.73TEE22 pKa = 4.58PEE24 pKa = 4.09DD25 pKa = 3.48PKK27 pKa = 10.94QEE29 pKa = 4.02QEE31 pKa = 4.25PEE33 pKa = 3.94QSTVTEE39 pKa = 4.0PVEE42 pKa = 3.96PAEE45 pKa = 4.49EE46 pKa = 4.64SPASTPAEE54 pKa = 4.22DD55 pKa = 5.0ADD57 pKa = 3.97AEE59 pKa = 4.48SAATEE64 pKa = 4.12PEE66 pKa = 4.02KK67 pKa = 10.57TEE69 pKa = 3.75QEE71 pKa = 4.15EE72 pKa = 4.46PEE74 pKa = 4.16EE75 pKa = 4.41SGEE78 pKa = 4.0KK79 pKa = 10.0PEE81 pKa = 4.19YY82 pKa = 10.21FFNGEE87 pKa = 4.03AVSVEE92 pKa = 4.04VPEE95 pKa = 4.28EE96 pKa = 3.63VRR98 pKa = 11.84NALKK102 pKa = 10.28EE103 pKa = 3.98AEE105 pKa = 4.12IDD107 pKa = 3.74EE108 pKa = 4.62KK109 pKa = 11.59ALLSQLFKK117 pKa = 10.97KK118 pKa = 10.75DD119 pKa = 3.18GDD121 pKa = 3.77FSLDD125 pKa = 3.12EE126 pKa = 4.0EE127 pKa = 4.82TRR129 pKa = 11.84TKK131 pKa = 10.96LEE133 pKa = 4.21SKK135 pKa = 10.39FGKK138 pKa = 8.56TLVDD142 pKa = 4.87GYY144 pKa = 11.49LNMYY148 pKa = 10.45KK149 pKa = 10.57GMNEE153 pKa = 3.91QAIKK157 pKa = 10.39AAAADD162 pKa = 3.97KK163 pKa = 9.71EE164 pKa = 4.37QQEE167 pKa = 4.71VTSKK171 pKa = 10.1QQAEE175 pKa = 4.21EE176 pKa = 4.1FMTTVGGEE184 pKa = 4.03EE185 pKa = 4.36GLVAMEE191 pKa = 5.82DD192 pKa = 4.36FILKK196 pKa = 10.28NFDD199 pKa = 3.74DD200 pKa = 4.43KK201 pKa = 11.56QIAAYY206 pKa = 8.87NAVMEE211 pKa = 4.7SEE213 pKa = 4.2SHH215 pKa = 5.2EE216 pKa = 4.04QQLLIIQTVKK226 pKa = 11.12AQMEE230 pKa = 4.37LSDD233 pKa = 4.3KK234 pKa = 10.6LQNGDD239 pKa = 4.13KK240 pKa = 10.54NIKK243 pKa = 10.25LLGDD247 pKa = 4.04DD248 pKa = 4.16TSSTPTGTPLDD259 pKa = 3.86KK260 pKa = 11.11GYY262 pKa = 8.86LTSDD266 pKa = 3.7EE267 pKa = 4.59FQAIMDD273 pKa = 3.96SDD275 pKa = 4.22KK276 pKa = 10.55YY277 pKa = 8.96WTDD280 pKa = 3.51FEE282 pKa = 4.7YY283 pKa = 9.48QTKK286 pKa = 10.19VDD288 pKa = 3.71AARR291 pKa = 11.84SAGLKK296 pKa = 10.43RR297 pKa = 11.84NLL299 pKa = 3.73
Molecular weight: 33.24 kDa
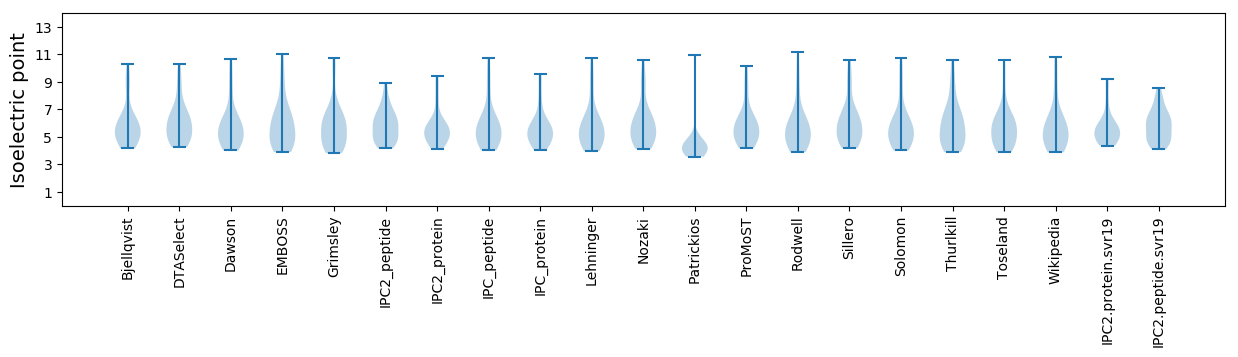
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9LQ90|A0A6B9LQ90_9CAUD Uncharacterized protein OS=Vibrio phage VP-HS15 OX=2686284 PE=4 SV=1
MM1 pKa = 7.4TEE3 pKa = 3.58LRR5 pKa = 11.84KK6 pKa = 10.08AISSRR11 pKa = 11.84AVKK14 pKa = 10.61HH15 pKa = 6.3KK16 pKa = 10.69DD17 pKa = 3.49NPDD20 pKa = 3.27NLPNIKK26 pKa = 10.08KK27 pKa = 10.1EE28 pKa = 4.07LIRR31 pKa = 11.84RR32 pKa = 11.84QNGKK36 pKa = 9.97CPITGRR42 pKa = 11.84DD43 pKa = 3.79LRR45 pKa = 11.84SMTSTNVVVDD55 pKa = 4.18HH56 pKa = 6.37DD57 pKa = 4.44HH58 pKa = 6.44KK59 pKa = 11.31SGVIRR64 pKa = 11.84AALPRR69 pKa = 11.84AINGLEE75 pKa = 3.97GKK77 pKa = 8.2LTNLCIRR84 pKa = 11.84WGGCTSKK91 pKa = 10.9RR92 pKa = 11.84DD93 pKa = 4.02IINMLRR99 pKa = 11.84GMADD103 pKa = 3.46YY104 pKa = 11.31LEE106 pKa = 4.38LHH108 pKa = 5.9QVPQTEE114 pKa = 4.78WIHH117 pKa = 6.23PEE119 pKa = 3.72HH120 pKa = 6.5LTPAEE125 pKa = 3.84ARR127 pKa = 11.84AKK129 pKa = 10.41RR130 pKa = 11.84NAKK133 pKa = 9.14ARR135 pKa = 11.84KK136 pKa = 9.25RR137 pKa = 11.84YY138 pKa = 9.41ALKK141 pKa = 10.79KK142 pKa = 9.56KK143 pKa = 10.69DD144 pKa = 3.37KK145 pKa = 10.92
MM1 pKa = 7.4TEE3 pKa = 3.58LRR5 pKa = 11.84KK6 pKa = 10.08AISSRR11 pKa = 11.84AVKK14 pKa = 10.61HH15 pKa = 6.3KK16 pKa = 10.69DD17 pKa = 3.49NPDD20 pKa = 3.27NLPNIKK26 pKa = 10.08KK27 pKa = 10.1EE28 pKa = 4.07LIRR31 pKa = 11.84RR32 pKa = 11.84QNGKK36 pKa = 9.97CPITGRR42 pKa = 11.84DD43 pKa = 3.79LRR45 pKa = 11.84SMTSTNVVVDD55 pKa = 4.18HH56 pKa = 6.37DD57 pKa = 4.44HH58 pKa = 6.44KK59 pKa = 11.31SGVIRR64 pKa = 11.84AALPRR69 pKa = 11.84AINGLEE75 pKa = 3.97GKK77 pKa = 8.2LTNLCIRR84 pKa = 11.84WGGCTSKK91 pKa = 10.9RR92 pKa = 11.84DD93 pKa = 4.02IINMLRR99 pKa = 11.84GMADD103 pKa = 3.46YY104 pKa = 11.31LEE106 pKa = 4.38LHH108 pKa = 5.9QVPQTEE114 pKa = 4.78WIHH117 pKa = 6.23PEE119 pKa = 3.72HH120 pKa = 6.5LTPAEE125 pKa = 3.84ARR127 pKa = 11.84AKK129 pKa = 10.41RR130 pKa = 11.84NAKK133 pKa = 9.14ARR135 pKa = 11.84KK136 pKa = 9.25RR137 pKa = 11.84YY138 pKa = 9.41ALKK141 pKa = 10.79KK142 pKa = 9.56KK143 pKa = 10.69DD144 pKa = 3.37KK145 pKa = 10.92
Molecular weight: 16.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12585 |
61 |
1229 |
419.5 |
47.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.24 ± 0.499 | 0.985 ± 0.161 |
6.667 ± 0.246 | 7.66 ± 0.407 |
3.949 ± 0.195 | 7.31 ± 0.446 |
1.859 ± 0.215 | 5.427 ± 0.223 |
6.643 ± 0.39 | 8.176 ± 0.316 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.773 ± 0.209 | 4.974 ± 0.215 |
3.639 ± 0.193 | 4.346 ± 0.269 |
5.189 ± 0.268 | 5.729 ± 0.359 |
5.356 ± 0.342 | 6.126 ± 0.345 |
1.375 ± 0.148 | 3.576 ± 0.182 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
