
Shahe isopoda virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.31
Get precalculated fractions of proteins
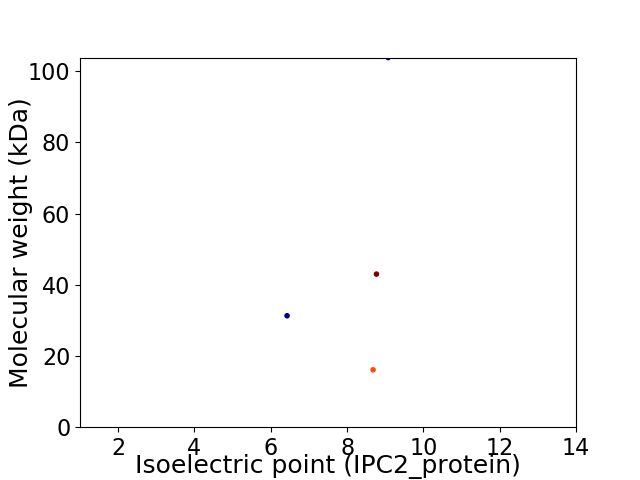
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A0A1L3KG25|A0A1L3KG25_9VIRU Uncharacterized protein OS=Shahe isopoda virus 5 OX=1923425 PE=4 SV=1
MM1 pKa = 7.41SLALLSLQDD10 pKa = 3.7PTFVAYY16 pKa = 9.53HH17 pKa = 6.63LCLISWDD24 pKa = 3.52GDD26 pKa = 3.57EE27 pKa = 4.07QGEE30 pKa = 4.57YY31 pKa = 10.96NFWEE35 pKa = 4.22WKK37 pKa = 9.49KK38 pKa = 10.81VYY40 pKa = 10.32GLEE43 pKa = 4.0KK44 pKa = 10.39HH45 pKa = 5.27VTAEE49 pKa = 4.26MVCDD53 pKa = 3.81ALEE56 pKa = 4.66GPGPQGEE63 pKa = 4.42ANEE66 pKa = 4.25EE67 pKa = 4.13SSALFGMQQIASFPAIRR84 pKa = 11.84AGLLEE89 pKa = 4.63KK90 pKa = 10.1KK91 pKa = 9.21WKK93 pKa = 9.38EE94 pKa = 3.76AGLAPPRR101 pKa = 11.84YY102 pKa = 8.13WHH104 pKa = 6.76LLTRR108 pKa = 11.84GGQQRR113 pKa = 11.84IITLTRR119 pKa = 11.84AIQEE123 pKa = 4.14RR124 pKa = 11.84FGTEE128 pKa = 3.05WRR130 pKa = 11.84GCSGTWASIGATFTLRR146 pKa = 11.84RR147 pKa = 11.84AEE149 pKa = 4.08QASQQLTVGDD159 pKa = 5.03LDD161 pKa = 4.31PEE163 pKa = 4.69STCLLWRR170 pKa = 11.84RR171 pKa = 11.84DD172 pKa = 3.06WQEE175 pKa = 4.09NYY177 pKa = 8.54WQVEE181 pKa = 4.1LFGPQPLGNRR191 pKa = 11.84AGGCRR196 pKa = 11.84FYY198 pKa = 11.74VEE200 pKa = 5.22GNTALQPGGAGASAAPTRR218 pKa = 11.84KK219 pKa = 9.42ARR221 pKa = 11.84RR222 pKa = 11.84GGKK225 pKa = 7.27EE226 pKa = 2.95ARR228 pKa = 11.84ARR230 pKa = 11.84HH231 pKa = 4.6NAKK234 pKa = 10.23RR235 pKa = 11.84FAEE238 pKa = 4.15KK239 pKa = 10.72LLLLEE244 pKa = 4.44SRR246 pKa = 11.84GEE248 pKa = 3.99LGSWANGWVFDD259 pKa = 4.19IPDD262 pKa = 3.89PVRR265 pKa = 11.84DD266 pKa = 3.37RR267 pKa = 11.84ARR269 pKa = 11.84QYY271 pKa = 11.0LPPDD275 pKa = 3.54FVIPP279 pKa = 4.07
MM1 pKa = 7.41SLALLSLQDD10 pKa = 3.7PTFVAYY16 pKa = 9.53HH17 pKa = 6.63LCLISWDD24 pKa = 3.52GDD26 pKa = 3.57EE27 pKa = 4.07QGEE30 pKa = 4.57YY31 pKa = 10.96NFWEE35 pKa = 4.22WKK37 pKa = 9.49KK38 pKa = 10.81VYY40 pKa = 10.32GLEE43 pKa = 4.0KK44 pKa = 10.39HH45 pKa = 5.27VTAEE49 pKa = 4.26MVCDD53 pKa = 3.81ALEE56 pKa = 4.66GPGPQGEE63 pKa = 4.42ANEE66 pKa = 4.25EE67 pKa = 4.13SSALFGMQQIASFPAIRR84 pKa = 11.84AGLLEE89 pKa = 4.63KK90 pKa = 10.1KK91 pKa = 9.21WKK93 pKa = 9.38EE94 pKa = 3.76AGLAPPRR101 pKa = 11.84YY102 pKa = 8.13WHH104 pKa = 6.76LLTRR108 pKa = 11.84GGQQRR113 pKa = 11.84IITLTRR119 pKa = 11.84AIQEE123 pKa = 4.14RR124 pKa = 11.84FGTEE128 pKa = 3.05WRR130 pKa = 11.84GCSGTWASIGATFTLRR146 pKa = 11.84RR147 pKa = 11.84AEE149 pKa = 4.08QASQQLTVGDD159 pKa = 5.03LDD161 pKa = 4.31PEE163 pKa = 4.69STCLLWRR170 pKa = 11.84RR171 pKa = 11.84DD172 pKa = 3.06WQEE175 pKa = 4.09NYY177 pKa = 8.54WQVEE181 pKa = 4.1LFGPQPLGNRR191 pKa = 11.84AGGCRR196 pKa = 11.84FYY198 pKa = 11.74VEE200 pKa = 5.22GNTALQPGGAGASAAPTRR218 pKa = 11.84KK219 pKa = 9.42ARR221 pKa = 11.84RR222 pKa = 11.84GGKK225 pKa = 7.27EE226 pKa = 2.95ARR228 pKa = 11.84ARR230 pKa = 11.84HH231 pKa = 4.6NAKK234 pKa = 10.23RR235 pKa = 11.84FAEE238 pKa = 4.15KK239 pKa = 10.72LLLLEE244 pKa = 4.44SRR246 pKa = 11.84GEE248 pKa = 3.99LGSWANGWVFDD259 pKa = 4.19IPDD262 pKa = 3.89PVRR265 pKa = 11.84DD266 pKa = 3.37RR267 pKa = 11.84ARR269 pKa = 11.84QYY271 pKa = 11.0LPPDD275 pKa = 3.54FVIPP279 pKa = 4.07
Molecular weight: 31.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG48|A0A1L3KG48_9VIRU RNA-directed RNA polymerase OS=Shahe isopoda virus 5 OX=1923425 PE=4 SV=1
MM1 pKa = 7.74AMNRR5 pKa = 11.84VNTTSGSGRR14 pKa = 11.84KK15 pKa = 8.12STVSRR20 pKa = 11.84STSRR24 pKa = 11.84PRR26 pKa = 11.84WSVMLSKK33 pKa = 11.07GPALKK38 pKa = 10.03EE39 pKa = 3.53RR40 pKa = 11.84RR41 pKa = 11.84TRR43 pKa = 11.84RR44 pKa = 11.84VQHH47 pKa = 6.47CSGCNRR53 pKa = 11.84SHH55 pKa = 7.4PFLPFEE61 pKa = 4.55PDD63 pKa = 2.89CLRR66 pKa = 11.84RR67 pKa = 11.84SGRR70 pKa = 11.84RR71 pKa = 11.84QDD73 pKa = 4.35LPPHH77 pKa = 5.79ATGTSLRR84 pKa = 11.84EE85 pKa = 3.72VASRR89 pKa = 11.84GSSRR93 pKa = 11.84SLGQSRR99 pKa = 11.84RR100 pKa = 11.84DD101 pKa = 3.5SVRR104 pKa = 11.84SGGVAAARR112 pKa = 11.84GPVLGQHH119 pKa = 5.94SRR121 pKa = 11.84SDD123 pKa = 3.99GLNKK127 pKa = 10.07LHH129 pKa = 6.69NSSRR133 pKa = 11.84SATSTQSLPAFSGEE147 pKa = 4.0GTGRR151 pKa = 11.84RR152 pKa = 11.84TTGRR156 pKa = 11.84LSSSGPSPWEE166 pKa = 3.8TEE168 pKa = 3.92RR169 pKa = 11.84VGAGFMWRR177 pKa = 11.84EE178 pKa = 4.07TPPSSRR184 pKa = 11.84VEE186 pKa = 4.0LGQALPQPAKK196 pKa = 10.49QGGEE200 pKa = 3.88EE201 pKa = 3.96KK202 pKa = 10.4RR203 pKa = 11.84RR204 pKa = 11.84ARR206 pKa = 11.84ATMPSGSRR214 pKa = 11.84KK215 pKa = 9.97SSFCSSRR222 pKa = 11.84EE223 pKa = 3.45EE224 pKa = 4.14SLAVGQTDD232 pKa = 3.98GSSTSPIQSVTEE244 pKa = 3.93QGSISPPISSSRR256 pKa = 11.84KK257 pKa = 7.23GGAYY261 pKa = 9.85AGPSRR266 pKa = 11.84GEE268 pKa = 4.13GPAPEE273 pKa = 4.32HH274 pKa = 6.06EE275 pKa = 4.71KK276 pKa = 10.45PVKK279 pKa = 10.27AKK281 pKa = 10.38CDD283 pKa = 3.42EE284 pKa = 4.28TFVPDD289 pKa = 3.23RR290 pKa = 11.84RR291 pKa = 11.84VRR293 pKa = 11.84ILGNKK298 pKa = 8.58NLVGRR303 pKa = 11.84GARR306 pKa = 11.84KK307 pKa = 9.7KK308 pKa = 9.81IRR310 pKa = 11.84RR311 pKa = 11.84EE312 pKa = 4.06VPCVDD317 pKa = 5.22DD318 pKa = 4.3EE319 pKa = 5.0LLGHH323 pKa = 7.14LLRR326 pKa = 11.84KK327 pKa = 9.16FAFVPRR333 pKa = 11.84TAEE336 pKa = 3.78LMVVMSRR343 pKa = 11.84TAQAFYY349 pKa = 10.85KK350 pKa = 10.82GFDD353 pKa = 3.38LSEE356 pKa = 3.89WTEE359 pKa = 4.0LEE361 pKa = 4.24LYY363 pKa = 7.86KK364 pKa = 8.86TTVMTITAAMDD375 pKa = 3.41IPDD378 pKa = 3.89EE379 pKa = 4.35EE380 pKa = 4.56EE381 pKa = 3.8ACRR384 pKa = 11.84QHH386 pKa = 6.85LKK388 pKa = 10.69SKK390 pKa = 10.67SLGEE394 pKa = 3.88ARR396 pKa = 11.84EE397 pKa = 4.11AHH399 pKa = 5.61AKK401 pKa = 8.21MMKK404 pKa = 9.96GHH406 pKa = 7.57LGNVRR411 pKa = 11.84NIFGVGRR418 pKa = 11.84TAVLPGKK425 pKa = 10.75GSGXDD430 pKa = 3.44RR431 pKa = 11.84LRR433 pKa = 11.84ITAHH437 pKa = 5.67CQRR440 pKa = 11.84EE441 pKa = 3.87VDD443 pKa = 3.81LTNNRR448 pKa = 11.84LPGCVSRR455 pKa = 11.84AGPQPCATKK464 pKa = 10.41RR465 pKa = 11.84STSRR469 pKa = 11.84LVSPVEE475 pKa = 3.65GWEE478 pKa = 4.31GVIVTHH484 pKa = 6.46SDD486 pKa = 3.78CVCNEE491 pKa = 3.26MRR493 pKa = 11.84ALHH496 pKa = 6.37HH497 pKa = 6.43RR498 pKa = 11.84HH499 pKa = 5.69QKK501 pKa = 7.58TAPAPTRR508 pKa = 11.84EE509 pKa = 4.03GLAMLKK515 pKa = 10.38KK516 pKa = 9.89KK517 pKa = 9.16VHH519 pKa = 5.79EE520 pKa = 4.26LFVVEE525 pKa = 4.72GVPGVIPKK533 pKa = 9.68PRR535 pKa = 11.84EE536 pKa = 3.82AVLEE540 pKa = 4.5HH541 pKa = 6.04YY542 pKa = 10.32SGRR545 pKa = 11.84QLAEE549 pKa = 3.81FTRR552 pKa = 11.84ALRR555 pKa = 11.84SLGEE559 pKa = 4.13RR560 pKa = 11.84PVCARR565 pKa = 11.84DD566 pKa = 3.34ADD568 pKa = 3.87VKK570 pKa = 10.9MFLKK574 pKa = 10.38DD575 pKa = 3.43DD576 pKa = 4.08KK577 pKa = 11.19YY578 pKa = 10.74VPEE581 pKa = 4.09YY582 pKa = 10.94AQIKK586 pKa = 9.77APRR589 pKa = 11.84CIQYY593 pKa = 10.08RR594 pKa = 11.84DD595 pKa = 3.1KK596 pKa = 10.97RR597 pKa = 11.84YY598 pKa = 9.53CLEE601 pKa = 3.92LARR604 pKa = 11.84YY605 pKa = 7.96LQIIEE610 pKa = 3.9GRR612 pKa = 11.84VYY614 pKa = 10.54GAEE617 pKa = 3.89DD618 pKa = 3.57AFGHH622 pKa = 6.12RR623 pKa = 11.84LIAKK627 pKa = 8.53GRR629 pKa = 11.84NLAQRR634 pKa = 11.84GADD637 pKa = 3.17LWAKK641 pKa = 10.47ANEE644 pKa = 4.21FADD647 pKa = 4.08PLFLLLDD654 pKa = 4.15ASNFDD659 pKa = 3.29AHH661 pKa = 6.56VATGLLKK668 pKa = 10.0IEE670 pKa = 4.14HH671 pKa = 5.8STYY674 pKa = 10.66IKK676 pKa = 9.43NTKK679 pKa = 9.69KK680 pKa = 10.39SARR683 pKa = 11.84HH684 pKa = 4.26MLRR687 pKa = 11.84WLLKK691 pKa = 8.74QQLINRR697 pKa = 11.84GRR699 pKa = 11.84TKK701 pKa = 10.94NGTTYY706 pKa = 9.14LTPGTRR712 pKa = 11.84MSGDD716 pKa = 3.36MNTGLGNSILMAGMLEE732 pKa = 4.72CYY734 pKa = 10.12LEE736 pKa = 4.13KK737 pKa = 10.79CGIKK741 pKa = 10.31GAVYY745 pKa = 10.76VDD747 pKa = 4.03GDD749 pKa = 3.83DD750 pKa = 3.56SVVVVEE756 pKa = 5.08RR757 pKa = 11.84QQQHH761 pKa = 6.0KK762 pKa = 9.22LLPVAPFFLQFGMEE776 pKa = 4.15MKK778 pKa = 10.87YY779 pKa = 10.17EE780 pKa = 4.05ATNEE784 pKa = 3.58FSQVDD789 pKa = 4.04FCQCRR794 pKa = 11.84PVQVDD799 pKa = 3.76GKK801 pKa = 9.14WVLSRR806 pKa = 11.84DD807 pKa = 3.7PKK809 pKa = 10.67RR810 pKa = 11.84VLTRR814 pKa = 11.84PLWTTRR820 pKa = 11.84EE821 pKa = 4.11MGDD824 pKa = 3.48KK825 pKa = 10.75LAGRR829 pKa = 11.84YY830 pKa = 9.16LKK832 pKa = 10.93GLGLGEE838 pKa = 4.24IAVNWGLPLGSVLGARR854 pKa = 11.84LYY856 pKa = 10.7EE857 pKa = 4.74IGEE860 pKa = 4.28GKK862 pKa = 8.35PWSYY866 pKa = 11.01EE867 pKa = 3.47FHH869 pKa = 7.22PGMKK873 pKa = 9.6ARR875 pKa = 11.84EE876 pKa = 4.18YY877 pKa = 11.42GRR879 pKa = 11.84IDD881 pKa = 3.35TPQPSWATRR890 pKa = 11.84MSFFEE895 pKa = 3.83AWGISPEE902 pKa = 3.98EE903 pKa = 3.73QEE905 pKa = 4.42AIEE908 pKa = 3.9RR909 pKa = 11.84SIRR912 pKa = 11.84SIHH915 pKa = 5.8RR916 pKa = 11.84TPTLPQDD923 pKa = 3.95LLEE926 pKa = 5.12GDD928 pKa = 3.84TLGPGFDD935 pKa = 3.5RR936 pKa = 11.84RR937 pKa = 3.77
MM1 pKa = 7.74AMNRR5 pKa = 11.84VNTTSGSGRR14 pKa = 11.84KK15 pKa = 8.12STVSRR20 pKa = 11.84STSRR24 pKa = 11.84PRR26 pKa = 11.84WSVMLSKK33 pKa = 11.07GPALKK38 pKa = 10.03EE39 pKa = 3.53RR40 pKa = 11.84RR41 pKa = 11.84TRR43 pKa = 11.84RR44 pKa = 11.84VQHH47 pKa = 6.47CSGCNRR53 pKa = 11.84SHH55 pKa = 7.4PFLPFEE61 pKa = 4.55PDD63 pKa = 2.89CLRR66 pKa = 11.84RR67 pKa = 11.84SGRR70 pKa = 11.84RR71 pKa = 11.84QDD73 pKa = 4.35LPPHH77 pKa = 5.79ATGTSLRR84 pKa = 11.84EE85 pKa = 3.72VASRR89 pKa = 11.84GSSRR93 pKa = 11.84SLGQSRR99 pKa = 11.84RR100 pKa = 11.84DD101 pKa = 3.5SVRR104 pKa = 11.84SGGVAAARR112 pKa = 11.84GPVLGQHH119 pKa = 5.94SRR121 pKa = 11.84SDD123 pKa = 3.99GLNKK127 pKa = 10.07LHH129 pKa = 6.69NSSRR133 pKa = 11.84SATSTQSLPAFSGEE147 pKa = 4.0GTGRR151 pKa = 11.84RR152 pKa = 11.84TTGRR156 pKa = 11.84LSSSGPSPWEE166 pKa = 3.8TEE168 pKa = 3.92RR169 pKa = 11.84VGAGFMWRR177 pKa = 11.84EE178 pKa = 4.07TPPSSRR184 pKa = 11.84VEE186 pKa = 4.0LGQALPQPAKK196 pKa = 10.49QGGEE200 pKa = 3.88EE201 pKa = 3.96KK202 pKa = 10.4RR203 pKa = 11.84RR204 pKa = 11.84ARR206 pKa = 11.84ATMPSGSRR214 pKa = 11.84KK215 pKa = 9.97SSFCSSRR222 pKa = 11.84EE223 pKa = 3.45EE224 pKa = 4.14SLAVGQTDD232 pKa = 3.98GSSTSPIQSVTEE244 pKa = 3.93QGSISPPISSSRR256 pKa = 11.84KK257 pKa = 7.23GGAYY261 pKa = 9.85AGPSRR266 pKa = 11.84GEE268 pKa = 4.13GPAPEE273 pKa = 4.32HH274 pKa = 6.06EE275 pKa = 4.71KK276 pKa = 10.45PVKK279 pKa = 10.27AKK281 pKa = 10.38CDD283 pKa = 3.42EE284 pKa = 4.28TFVPDD289 pKa = 3.23RR290 pKa = 11.84RR291 pKa = 11.84VRR293 pKa = 11.84ILGNKK298 pKa = 8.58NLVGRR303 pKa = 11.84GARR306 pKa = 11.84KK307 pKa = 9.7KK308 pKa = 9.81IRR310 pKa = 11.84RR311 pKa = 11.84EE312 pKa = 4.06VPCVDD317 pKa = 5.22DD318 pKa = 4.3EE319 pKa = 5.0LLGHH323 pKa = 7.14LLRR326 pKa = 11.84KK327 pKa = 9.16FAFVPRR333 pKa = 11.84TAEE336 pKa = 3.78LMVVMSRR343 pKa = 11.84TAQAFYY349 pKa = 10.85KK350 pKa = 10.82GFDD353 pKa = 3.38LSEE356 pKa = 3.89WTEE359 pKa = 4.0LEE361 pKa = 4.24LYY363 pKa = 7.86KK364 pKa = 8.86TTVMTITAAMDD375 pKa = 3.41IPDD378 pKa = 3.89EE379 pKa = 4.35EE380 pKa = 4.56EE381 pKa = 3.8ACRR384 pKa = 11.84QHH386 pKa = 6.85LKK388 pKa = 10.69SKK390 pKa = 10.67SLGEE394 pKa = 3.88ARR396 pKa = 11.84EE397 pKa = 4.11AHH399 pKa = 5.61AKK401 pKa = 8.21MMKK404 pKa = 9.96GHH406 pKa = 7.57LGNVRR411 pKa = 11.84NIFGVGRR418 pKa = 11.84TAVLPGKK425 pKa = 10.75GSGXDD430 pKa = 3.44RR431 pKa = 11.84LRR433 pKa = 11.84ITAHH437 pKa = 5.67CQRR440 pKa = 11.84EE441 pKa = 3.87VDD443 pKa = 3.81LTNNRR448 pKa = 11.84LPGCVSRR455 pKa = 11.84AGPQPCATKK464 pKa = 10.41RR465 pKa = 11.84STSRR469 pKa = 11.84LVSPVEE475 pKa = 3.65GWEE478 pKa = 4.31GVIVTHH484 pKa = 6.46SDD486 pKa = 3.78CVCNEE491 pKa = 3.26MRR493 pKa = 11.84ALHH496 pKa = 6.37HH497 pKa = 6.43RR498 pKa = 11.84HH499 pKa = 5.69QKK501 pKa = 7.58TAPAPTRR508 pKa = 11.84EE509 pKa = 4.03GLAMLKK515 pKa = 10.38KK516 pKa = 9.89KK517 pKa = 9.16VHH519 pKa = 5.79EE520 pKa = 4.26LFVVEE525 pKa = 4.72GVPGVIPKK533 pKa = 9.68PRR535 pKa = 11.84EE536 pKa = 3.82AVLEE540 pKa = 4.5HH541 pKa = 6.04YY542 pKa = 10.32SGRR545 pKa = 11.84QLAEE549 pKa = 3.81FTRR552 pKa = 11.84ALRR555 pKa = 11.84SLGEE559 pKa = 4.13RR560 pKa = 11.84PVCARR565 pKa = 11.84DD566 pKa = 3.34ADD568 pKa = 3.87VKK570 pKa = 10.9MFLKK574 pKa = 10.38DD575 pKa = 3.43DD576 pKa = 4.08KK577 pKa = 11.19YY578 pKa = 10.74VPEE581 pKa = 4.09YY582 pKa = 10.94AQIKK586 pKa = 9.77APRR589 pKa = 11.84CIQYY593 pKa = 10.08RR594 pKa = 11.84DD595 pKa = 3.1KK596 pKa = 10.97RR597 pKa = 11.84YY598 pKa = 9.53CLEE601 pKa = 3.92LARR604 pKa = 11.84YY605 pKa = 7.96LQIIEE610 pKa = 3.9GRR612 pKa = 11.84VYY614 pKa = 10.54GAEE617 pKa = 3.89DD618 pKa = 3.57AFGHH622 pKa = 6.12RR623 pKa = 11.84LIAKK627 pKa = 8.53GRR629 pKa = 11.84NLAQRR634 pKa = 11.84GADD637 pKa = 3.17LWAKK641 pKa = 10.47ANEE644 pKa = 4.21FADD647 pKa = 4.08PLFLLLDD654 pKa = 4.15ASNFDD659 pKa = 3.29AHH661 pKa = 6.56VATGLLKK668 pKa = 10.0IEE670 pKa = 4.14HH671 pKa = 5.8STYY674 pKa = 10.66IKK676 pKa = 9.43NTKK679 pKa = 9.69KK680 pKa = 10.39SARR683 pKa = 11.84HH684 pKa = 4.26MLRR687 pKa = 11.84WLLKK691 pKa = 8.74QQLINRR697 pKa = 11.84GRR699 pKa = 11.84TKK701 pKa = 10.94NGTTYY706 pKa = 9.14LTPGTRR712 pKa = 11.84MSGDD716 pKa = 3.36MNTGLGNSILMAGMLEE732 pKa = 4.72CYY734 pKa = 10.12LEE736 pKa = 4.13KK737 pKa = 10.79CGIKK741 pKa = 10.31GAVYY745 pKa = 10.76VDD747 pKa = 4.03GDD749 pKa = 3.83DD750 pKa = 3.56SVVVVEE756 pKa = 5.08RR757 pKa = 11.84QQQHH761 pKa = 6.0KK762 pKa = 9.22LLPVAPFFLQFGMEE776 pKa = 4.15MKK778 pKa = 10.87YY779 pKa = 10.17EE780 pKa = 4.05ATNEE784 pKa = 3.58FSQVDD789 pKa = 4.04FCQCRR794 pKa = 11.84PVQVDD799 pKa = 3.76GKK801 pKa = 9.14WVLSRR806 pKa = 11.84DD807 pKa = 3.7PKK809 pKa = 10.67RR810 pKa = 11.84VLTRR814 pKa = 11.84PLWTTRR820 pKa = 11.84EE821 pKa = 4.11MGDD824 pKa = 3.48KK825 pKa = 10.75LAGRR829 pKa = 11.84YY830 pKa = 9.16LKK832 pKa = 10.93GLGLGEE838 pKa = 4.24IAVNWGLPLGSVLGARR854 pKa = 11.84LYY856 pKa = 10.7EE857 pKa = 4.74IGEE860 pKa = 4.28GKK862 pKa = 8.35PWSYY866 pKa = 11.01EE867 pKa = 3.47FHH869 pKa = 7.22PGMKK873 pKa = 9.6ARR875 pKa = 11.84EE876 pKa = 4.18YY877 pKa = 11.42GRR879 pKa = 11.84IDD881 pKa = 3.35TPQPSWATRR890 pKa = 11.84MSFFEE895 pKa = 3.83AWGISPEE902 pKa = 3.98EE903 pKa = 3.73QEE905 pKa = 4.42AIEE908 pKa = 3.9RR909 pKa = 11.84SIRR912 pKa = 11.84SIHH915 pKa = 5.8RR916 pKa = 11.84TPTLPQDD923 pKa = 3.95LLEE926 pKa = 5.12GDD928 pKa = 3.84TLGPGFDD935 pKa = 3.5RR936 pKa = 11.84RR937 pKa = 3.77
Molecular weight: 103.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1765 |
149 |
937 |
441.3 |
48.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.215 ± 0.832 | 1.7 ± 0.265 |
3.739 ± 0.337 | 5.949 ± 1.054 |
3.286 ± 0.561 | 9.292 ± 0.567 |
1.813 ± 0.597 | 3.626 ± 0.552 |
4.816 ± 0.505 | 8.952 ± 1.62 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.493 ± 0.348 | 2.89 ± 0.596 |
6.516 ± 0.638 | 4.306 ± 0.603 |
8.272 ± 1.488 | 7.875 ± 1.071 |
6.346 ± 0.861 | 5.722 ± 0.501 |
1.7 ± 0.618 | 2.436 ± 0.603 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
