
Drosophila subobscura Nora virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
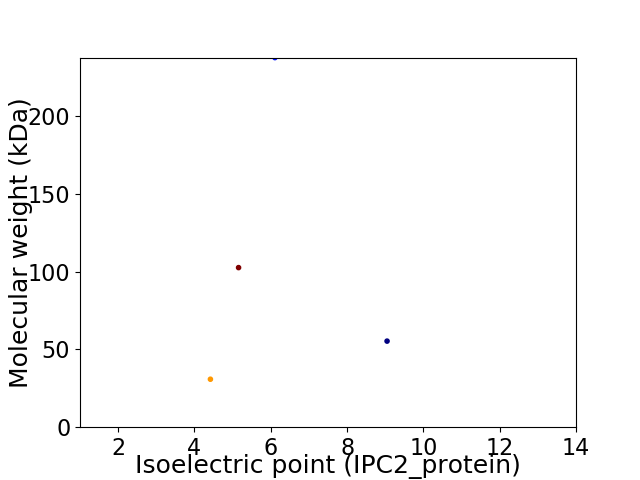
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075E1R9|A0A075E1R9_9VIRU VP2 OS=Drosophila subobscura Nora virus OX=1500865 GN=VP2 PE=4 SV=1
MM1 pKa = 7.49ALKK4 pKa = 10.69EE5 pKa = 4.29EE6 pKa = 4.59IFDD9 pKa = 3.82QNTTLFSVLDD19 pKa = 4.05EE20 pKa = 4.64NEE22 pKa = 3.98VTEE25 pKa = 4.5TKK27 pKa = 10.4QIQNSVTTVQTQIDD41 pKa = 3.98QQKK44 pKa = 9.99LQLDD48 pKa = 4.31GLAKK52 pKa = 10.62VVDD55 pKa = 4.11QNQSRR60 pKa = 11.84NEE62 pKa = 3.87EE63 pKa = 3.82QFVNINTIIVGMNDD77 pKa = 5.1DD78 pKa = 3.78IDD80 pKa = 3.87KK81 pKa = 11.43LKK83 pKa = 9.45ITTTNLVQQTNSLNSSVAEE102 pKa = 4.11LEE104 pKa = 4.39TFVPRR109 pKa = 11.84VNSLEE114 pKa = 4.12VQTDD118 pKa = 3.42ANTKK122 pKa = 10.31SISALEE128 pKa = 4.1GTTDD132 pKa = 5.46GIQDD136 pKa = 4.41SINTINDD143 pKa = 4.51GIVEE147 pKa = 4.29LKK149 pKa = 10.63DD150 pKa = 3.68DD151 pKa = 4.37NKK153 pKa = 11.18DD154 pKa = 3.34LSSQIQALDD163 pKa = 3.32SRR165 pKa = 11.84ITILEE170 pKa = 4.11SKK172 pKa = 9.7IKK174 pKa = 10.63SSFIEE179 pKa = 4.01KK180 pKa = 9.9SVQYY184 pKa = 10.3KK185 pKa = 10.23ISYY188 pKa = 9.74SNNSDD193 pKa = 3.29VQRR196 pKa = 11.84VLNFYY201 pKa = 11.14ALGPVKK207 pKa = 10.35YY208 pKa = 8.78GQSFSVMQTPTSTFSTTLTINRR230 pKa = 11.84PYY232 pKa = 9.62PQLSAITYY240 pKa = 8.04PGVPVLIGMQVSLSGTYY257 pKa = 10.72PMGTNVFEE265 pKa = 4.35SLKK268 pKa = 10.46DD269 pKa = 3.56RR270 pKa = 11.84PAIIYY275 pKa = 9.95SEE277 pKa = 4.13QQ278 pKa = 3.08
MM1 pKa = 7.49ALKK4 pKa = 10.69EE5 pKa = 4.29EE6 pKa = 4.59IFDD9 pKa = 3.82QNTTLFSVLDD19 pKa = 4.05EE20 pKa = 4.64NEE22 pKa = 3.98VTEE25 pKa = 4.5TKK27 pKa = 10.4QIQNSVTTVQTQIDD41 pKa = 3.98QQKK44 pKa = 9.99LQLDD48 pKa = 4.31GLAKK52 pKa = 10.62VVDD55 pKa = 4.11QNQSRR60 pKa = 11.84NEE62 pKa = 3.87EE63 pKa = 3.82QFVNINTIIVGMNDD77 pKa = 5.1DD78 pKa = 3.78IDD80 pKa = 3.87KK81 pKa = 11.43LKK83 pKa = 9.45ITTTNLVQQTNSLNSSVAEE102 pKa = 4.11LEE104 pKa = 4.39TFVPRR109 pKa = 11.84VNSLEE114 pKa = 4.12VQTDD118 pKa = 3.42ANTKK122 pKa = 10.31SISALEE128 pKa = 4.1GTTDD132 pKa = 5.46GIQDD136 pKa = 4.41SINTINDD143 pKa = 4.51GIVEE147 pKa = 4.29LKK149 pKa = 10.63DD150 pKa = 3.68DD151 pKa = 4.37NKK153 pKa = 11.18DD154 pKa = 3.34LSSQIQALDD163 pKa = 3.32SRR165 pKa = 11.84ITILEE170 pKa = 4.11SKK172 pKa = 9.7IKK174 pKa = 10.63SSFIEE179 pKa = 4.01KK180 pKa = 9.9SVQYY184 pKa = 10.3KK185 pKa = 10.23ISYY188 pKa = 9.74SNNSDD193 pKa = 3.29VQRR196 pKa = 11.84VLNFYY201 pKa = 11.14ALGPVKK207 pKa = 10.35YY208 pKa = 8.78GQSFSVMQTPTSTFSTTLTINRR230 pKa = 11.84PYY232 pKa = 9.62PQLSAITYY240 pKa = 8.04PGVPVLIGMQVSLSGTYY257 pKa = 10.72PMGTNVFEE265 pKa = 4.35SLKK268 pKa = 10.46DD269 pKa = 3.56RR270 pKa = 11.84PAIIYY275 pKa = 9.95SEE277 pKa = 4.13QQ278 pKa = 3.08
Molecular weight: 30.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075E1L1|A0A075E1L1_9VIRU VP3 OS=Drosophila subobscura Nora virus OX=1500865 GN=VP3 PE=4 SV=1
MM1 pKa = 7.18STNKK5 pKa = 9.05TNKK8 pKa = 9.71KK9 pKa = 8.79GAQLEE14 pKa = 4.25RR15 pKa = 11.84VQIASTQVVDD25 pKa = 3.63TRR27 pKa = 11.84SKK29 pKa = 10.05RR30 pKa = 11.84RR31 pKa = 11.84QRR33 pKa = 11.84GTKK36 pKa = 9.92LDD38 pKa = 2.98IDD40 pKa = 4.06YY41 pKa = 9.18TVKK44 pKa = 10.71KK45 pKa = 10.54NNAEE49 pKa = 4.08KK50 pKa = 10.15EE51 pKa = 4.33QKK53 pKa = 10.54FLDD56 pKa = 3.97TEE58 pKa = 4.45VVDD61 pKa = 4.16TKK63 pKa = 11.09LDD65 pKa = 3.59KK66 pKa = 11.04QILYY70 pKa = 9.62EE71 pKa = 3.94KK72 pKa = 9.68RR73 pKa = 11.84QNHH76 pKa = 5.25TFIKK80 pKa = 9.66PKK82 pKa = 10.17LSMVSRR88 pKa = 11.84EE89 pKa = 4.03DD90 pKa = 3.85KK91 pKa = 10.35IVKK94 pKa = 10.32NKK96 pKa = 8.86VLRR99 pKa = 11.84GNEE102 pKa = 3.57RR103 pKa = 11.84ASAYY107 pKa = 10.06KK108 pKa = 10.25FMKK111 pKa = 10.5EE112 pKa = 3.76MVNSNKK118 pKa = 9.24VQSSWNVEE126 pKa = 3.87RR127 pKa = 11.84VKK129 pKa = 10.93EE130 pKa = 3.96VDD132 pKa = 3.36EE133 pKa = 4.3VDD135 pKa = 3.57LFFRR139 pKa = 11.84KK140 pKa = 9.83KK141 pKa = 8.26KK142 pKa = 8.58TKK144 pKa = 9.41PFSGFSIGEE153 pKa = 4.07LRR155 pKa = 11.84DD156 pKa = 3.76SLIVPSDD163 pKa = 3.89DD164 pKa = 3.86KK165 pKa = 11.67NIVPPIVMSSTNEE178 pKa = 3.6IITPSEE184 pKa = 4.34EE185 pKa = 3.85ISVSAILKK193 pKa = 9.72QLAALTAKK201 pKa = 10.12LDD203 pKa = 4.13KK204 pKa = 10.56IEE206 pKa = 4.28KK207 pKa = 9.91QNEE210 pKa = 3.76EE211 pKa = 4.52LIKK214 pKa = 10.81EE215 pKa = 4.19NNNLKK220 pKa = 9.3MEE222 pKa = 4.56RR223 pKa = 11.84EE224 pKa = 3.82QFLEE228 pKa = 4.08NVHH231 pKa = 7.1KK232 pKa = 10.42EE233 pKa = 4.11VEE235 pKa = 4.48LEE237 pKa = 4.17TKK239 pKa = 9.76PQEE242 pKa = 4.34KK243 pKa = 8.55KK244 pKa = 9.12TLNTKK249 pKa = 8.39KK250 pKa = 8.22TQQKK254 pKa = 10.42SLGVNLKK261 pKa = 8.34ITKK264 pKa = 9.11TKK266 pKa = 10.4IIGQEE271 pKa = 3.45EE272 pKa = 4.47SLQQKK277 pKa = 9.38FVDD280 pKa = 4.62KK281 pKa = 10.42PNKK284 pKa = 8.67PLKK287 pKa = 9.55PNKK290 pKa = 10.09NMLGDD295 pKa = 3.78KK296 pKa = 9.95LKK298 pKa = 11.12KK299 pKa = 10.28NIRR302 pKa = 11.84EE303 pKa = 4.21WYY305 pKa = 10.16NFDD308 pKa = 3.6SSKK311 pKa = 11.2LEE313 pKa = 3.81QHH315 pKa = 6.07KK316 pKa = 11.05KK317 pKa = 9.72EE318 pKa = 4.11VLSSVITNATFAEE331 pKa = 5.01KK332 pKa = 9.88IRR334 pKa = 11.84EE335 pKa = 4.16TGVPKK340 pKa = 10.57QKK342 pKa = 9.87IRR344 pKa = 11.84YY345 pKa = 5.68TSPVTLKK352 pKa = 10.16EE353 pKa = 4.21DD354 pKa = 3.39KK355 pKa = 10.86KK356 pKa = 10.51SIHH359 pKa = 6.36YY360 pKa = 9.85YY361 pKa = 9.75GYY363 pKa = 10.21KK364 pKa = 10.01PSGIPNKK371 pKa = 9.73VWWHH375 pKa = 4.88WVTTVTSVEE384 pKa = 4.13AYY386 pKa = 8.83EE387 pKa = 3.93KK388 pKa = 11.1AEE390 pKa = 3.76RR391 pKa = 11.84FLYY394 pKa = 10.39NQFKK398 pKa = 10.84RR399 pKa = 11.84EE400 pKa = 3.76MFIYY404 pKa = 7.93RR405 pKa = 11.84TKK407 pKa = 8.26WVKK410 pKa = 10.05YY411 pKa = 8.57SKK413 pKa = 10.56EE414 pKa = 3.83FNPYY418 pKa = 9.47LSEE421 pKa = 4.12PKK423 pKa = 9.38MVWMEE428 pKa = 3.95HH429 pKa = 5.66TSDD432 pKa = 3.53FDD434 pKa = 3.95IDD436 pKa = 3.86VSVAYY441 pKa = 10.26AFILKK446 pKa = 8.4WRR448 pKa = 11.84KK449 pKa = 8.7LVQTFKK455 pKa = 10.66PNKK458 pKa = 8.72PINSDD463 pKa = 3.1WYY465 pKa = 11.0KK466 pKa = 10.17STQQQQ471 pKa = 2.91
MM1 pKa = 7.18STNKK5 pKa = 9.05TNKK8 pKa = 9.71KK9 pKa = 8.79GAQLEE14 pKa = 4.25RR15 pKa = 11.84VQIASTQVVDD25 pKa = 3.63TRR27 pKa = 11.84SKK29 pKa = 10.05RR30 pKa = 11.84RR31 pKa = 11.84QRR33 pKa = 11.84GTKK36 pKa = 9.92LDD38 pKa = 2.98IDD40 pKa = 4.06YY41 pKa = 9.18TVKK44 pKa = 10.71KK45 pKa = 10.54NNAEE49 pKa = 4.08KK50 pKa = 10.15EE51 pKa = 4.33QKK53 pKa = 10.54FLDD56 pKa = 3.97TEE58 pKa = 4.45VVDD61 pKa = 4.16TKK63 pKa = 11.09LDD65 pKa = 3.59KK66 pKa = 11.04QILYY70 pKa = 9.62EE71 pKa = 3.94KK72 pKa = 9.68RR73 pKa = 11.84QNHH76 pKa = 5.25TFIKK80 pKa = 9.66PKK82 pKa = 10.17LSMVSRR88 pKa = 11.84EE89 pKa = 4.03DD90 pKa = 3.85KK91 pKa = 10.35IVKK94 pKa = 10.32NKK96 pKa = 8.86VLRR99 pKa = 11.84GNEE102 pKa = 3.57RR103 pKa = 11.84ASAYY107 pKa = 10.06KK108 pKa = 10.25FMKK111 pKa = 10.5EE112 pKa = 3.76MVNSNKK118 pKa = 9.24VQSSWNVEE126 pKa = 3.87RR127 pKa = 11.84VKK129 pKa = 10.93EE130 pKa = 3.96VDD132 pKa = 3.36EE133 pKa = 4.3VDD135 pKa = 3.57LFFRR139 pKa = 11.84KK140 pKa = 9.83KK141 pKa = 8.26KK142 pKa = 8.58TKK144 pKa = 9.41PFSGFSIGEE153 pKa = 4.07LRR155 pKa = 11.84DD156 pKa = 3.76SLIVPSDD163 pKa = 3.89DD164 pKa = 3.86KK165 pKa = 11.67NIVPPIVMSSTNEE178 pKa = 3.6IITPSEE184 pKa = 4.34EE185 pKa = 3.85ISVSAILKK193 pKa = 9.72QLAALTAKK201 pKa = 10.12LDD203 pKa = 4.13KK204 pKa = 10.56IEE206 pKa = 4.28KK207 pKa = 9.91QNEE210 pKa = 3.76EE211 pKa = 4.52LIKK214 pKa = 10.81EE215 pKa = 4.19NNNLKK220 pKa = 9.3MEE222 pKa = 4.56RR223 pKa = 11.84EE224 pKa = 3.82QFLEE228 pKa = 4.08NVHH231 pKa = 7.1KK232 pKa = 10.42EE233 pKa = 4.11VEE235 pKa = 4.48LEE237 pKa = 4.17TKK239 pKa = 9.76PQEE242 pKa = 4.34KK243 pKa = 8.55KK244 pKa = 9.12TLNTKK249 pKa = 8.39KK250 pKa = 8.22TQQKK254 pKa = 10.42SLGVNLKK261 pKa = 8.34ITKK264 pKa = 9.11TKK266 pKa = 10.4IIGQEE271 pKa = 3.45EE272 pKa = 4.47SLQQKK277 pKa = 9.38FVDD280 pKa = 4.62KK281 pKa = 10.42PNKK284 pKa = 8.67PLKK287 pKa = 9.55PNKK290 pKa = 10.09NMLGDD295 pKa = 3.78KK296 pKa = 9.95LKK298 pKa = 11.12KK299 pKa = 10.28NIRR302 pKa = 11.84EE303 pKa = 4.21WYY305 pKa = 10.16NFDD308 pKa = 3.6SSKK311 pKa = 11.2LEE313 pKa = 3.81QHH315 pKa = 6.07KK316 pKa = 11.05KK317 pKa = 9.72EE318 pKa = 4.11VLSSVITNATFAEE331 pKa = 5.01KK332 pKa = 9.88IRR334 pKa = 11.84EE335 pKa = 4.16TGVPKK340 pKa = 10.57QKK342 pKa = 9.87IRR344 pKa = 11.84YY345 pKa = 5.68TSPVTLKK352 pKa = 10.16EE353 pKa = 4.21DD354 pKa = 3.39KK355 pKa = 10.86KK356 pKa = 10.51SIHH359 pKa = 6.36YY360 pKa = 9.85YY361 pKa = 9.75GYY363 pKa = 10.21KK364 pKa = 10.01PSGIPNKK371 pKa = 9.73VWWHH375 pKa = 4.88WVTTVTSVEE384 pKa = 4.13AYY386 pKa = 8.83EE387 pKa = 3.93KK388 pKa = 11.1AEE390 pKa = 3.76RR391 pKa = 11.84FLYY394 pKa = 10.39NQFKK398 pKa = 10.84RR399 pKa = 11.84EE400 pKa = 3.76MFIYY404 pKa = 7.93RR405 pKa = 11.84TKK407 pKa = 8.26WVKK410 pKa = 10.05YY411 pKa = 8.57SKK413 pKa = 10.56EE414 pKa = 3.83FNPYY418 pKa = 9.47LSEE421 pKa = 4.12PKK423 pKa = 9.38MVWMEE428 pKa = 3.95HH429 pKa = 5.66TSDD432 pKa = 3.53FDD434 pKa = 3.95IDD436 pKa = 3.86VSVAYY441 pKa = 10.26AFILKK446 pKa = 8.4WRR448 pKa = 11.84KK449 pKa = 8.7LVQTFKK455 pKa = 10.66PNKK458 pKa = 8.72PINSDD463 pKa = 3.1WYY465 pKa = 11.0KK466 pKa = 10.17STQQQQ471 pKa = 2.91
Molecular weight: 55.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3764 |
278 |
2081 |
941.0 |
106.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.526 ± 0.93 | 1.089 ± 0.475 |
5.367 ± 0.36 | 6.854 ± 0.551 |
3.905 ± 0.16 | 5.234 ± 0.856 |
1.674 ± 0.331 | 7.519 ± 0.395 |
7.625 ± 2.168 | 8.369 ± 0.598 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.129 | 5.712 ± 0.558 |
4.437 ± 1.059 | 4.888 ± 0.669 |
4.118 ± 0.332 | 6.509 ± 0.904 |
6.961 ± 0.535 | 7.067 ± 0.571 |
1.488 ± 0.317 | 3.294 ± 0.209 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
